Abstract
Proton magnetic resonance spectroscopy (1H-MRS) provides specific metabolic information not otherwise observable by any other imaging method. 1H-MRS of the brain at 3 T is a new tool in the modern neuroradiological armamentarium whose main advantages, with respect to the well-established and technologically advanced 1.5-T 1H-MRS, include a higher signal-to-noise ratio, with a consequent increase in spatial and temporal resolutions, and better spectral resolution. These advantages allow the acquisition of higher quality and more easily quantifiable spectra in smaller voxels and/or in shorter times, and increase the sensitivity in metabolite detection. However, these advantages may be hampered by intrinsic field-dependent technical issues, such as decreased T2 signal, chemical shift dispersion errors, J-modulation anomalies, increased magnetic susceptibility, eddy current artifacts, challenges in designing and obtaining appropriate radiofrequency coils, magnetic field instability and safety hazards. All these limitations have been tackled by manufacturers and researchers and have received one or more solutions. Furthermore, advanced 1H-MRS techniques, such as specific spectral editing, fast 1H-MRS imaging and diffusion tensor 1H-MRS imaging, have been successfully implemented at 3 T. However, easier and more robust implementations of these techniques are still needed before they can become more widely used and undertake most of the clinical and research 1H-MRS applications.
Similar content being viewed by others
Introduction
Proton magnetic resonance spectroscopy (1H-MRS) of the brain is a non-invasive technique that supplies information about the presence and levels of different low-molecular weight chemicals. The metabolites detectable with 1H-MRS include the prominent resonances of N-acetylaspartate (NAA), choline (Cho) and creatine (Cr), and a variety of other resonances that might or might not be evident depending on the type and quality of spectra as well as on the pathological condition [1–3]. Several metabolites, such as lactate in epilepsy or mitochondrial disorders [3], phenylalanine in phenylketonuria [4], guanidinoacetate in guanidinoacetate methyltransferase deficiency [5] and glycine in nonketotic hyperglycinemia [6], can be observed only when their concentrations are several times higher than normal.
1H-MRS has been utilized to identify and characterize the metabolic changes associated with many neurological disorders. The list is very long and includes brain tumors, degenerative diseases such as Alzheimer’s, Huntington’s and Parkinson’s diseases, cerebrovascular diseases, metabolic disorders such as adrenoleukodystrophy and Canavan’s disease, epilepsy, multiple sclerosis and systemic diseases such as hepatic and renal failure [1–3]. Rare diseases also studied include creatine deficiency syndrome [7], variant Creutzfeldt-Jakob Disease [8], pantothenate kinase-associated neurodegeneration [9] and Rasmussen’s encephalitis [10]. Most of these studies have been performed using devices operating at 1.5 T, which has been considered the standard field for years.
In the last decade, with the approval of the US Food and Drug Administration for clinical use, MR systems at 3 T are proliferating, particularly at research centers [11]. With respect to the well-established MR technique at 1.5 T, switching to a higher field brings several advantages, such as an increased signal-to-noise ratio, with consequent enhanced spatial and temporal resolutions, and better spectral resolution, but also many limitations, such as installation issues, higher acoustic noise, device compatibility, system inhomogeneity, eddy current artifacts, misregistration errors, J-modulation anomalies, magnetic field instability and safety restrictions. Some technical characteristics, such as changes in relaxation times, chemical shift and susceptibility, can have both benefits and disadvantages [11–15]. These limitations necessitating changes in technical devices and acquisition strategies have led to a debate about the usefulness of higher field strength in clinical settings [14–20]. However, the new generation of 3-T systems presents a number of fundamental technical differences with respect to the first generation, which have reduced the concerns about the limitations as well as the benefits of 3-T over 1.5-T systems and increased the penetration of 3-T scanners into the clinical setting [18–20]. Furthermore, adapting the imaging procedures to changes produced by the higher field allows obtaining images with quality and/or acquisition speed superior to 1.5 T [21, 22]. A number of recent studies have evidenced the advantages of 3 T over 1.5 T for both conventional MRI and MR applications limited by insufficient sensitivity, such as MR angiography, functional MRI and 1H-MRS [19, 20, 23, 24]. This review focuses on brain 1H-MRS at 3 T, illustrating the advantages, the strategies to overcome the limitations and the advanced techniques.
Advantages and disadvantages of 1H-MRS at 3 T
Signal-to-noise ratio
The intensity of the MR signal is correlated linearly with the strength of the static magnetic field. Thus, in theory, the signal-to-noise ratio (SNR) would double when moving from 1.5 T to 3 T, but in practice the improvement ranges only from 20% to 50% [25–28]. In effect, the SNR depends on several other variables, such as T1 and T2 relaxation times, type of sequence, number of signal averages, size of sample volume, radiofrequency (RF) effects and shimming efficacy. T1 relaxation times increase at 3 T, leading to increased signal saturation for a given repetition time, while T2 relaxation times decrease. Therefore, the theoretical doubling of SNR cannot be achieved, owing to the use of repetition times (TR) in the order of the T1 decay times (and not infinitely long) and echo times (TE) in the order of the metabolite T2 decay times (and not close to 0) [25]. Furthermore, SNR declines progressively with the increase of TE, passing from values ∼35% better than 1.5 T at TE 30 ms, to values less than 25% at TE 144 ms, and to equity at TE 288 ms [28].
One of the possible approaches to increase the SNR is the use of multiple receiver coils. A well-designed phased-array (PA) head coil, in fact, has sensitivity significantly superior to that of the birdcage-type volume coil, which is more widely used. The SNR increases with the number of coil elements, mainly for the regions next to the coil array and to a lesser extent for those towards the center of the imaged object [29]. The signals obtained from the different coil elements can be combined by several processing tools, including nonparametric, semiparametric and parametric ones, selected on the basis of available prior knowledge about the data [30]. A surface PA coil can provide approximately a ten-fold increase in SNR at the brain cortex as compared to a standard head coil [31].
Spatial and temporal resolution
The increased SNR associated with higher magnetic fields permits shorter imaging times for a given spatial resolution, higher resolution for a given imaging time or the combination of both. The advantages of shorter examination times for patients, radiologists, technicians and hospital administrators are obvious. Higher spatial resolution improves the matching of metabolic and anatomical information, enhancing the clinical value of 1H-MRS [11].
Spectra can be acquired from single-voxel, or from two-dimensional (2D) or three-dimensional (3D) multivoxels that are generally referred to as chemical shift imaging (CSI), spectroscopic imaging (SI) or MRS imaging (MRSI) (Fig. 1). At 1.5 T, most 1H-MRS studies have been performed with a spatial resolution of 1 cm3 or more, while the resolution of routine MRI is about 1 mm3 [32]. At 3 T, voxels well below 1 cm3 can be obtained with good SNR and acceptable acquisition times (Fig. 1) [25, 31, 32]. Using a birdcage head coil, 3D MRSI with 0.75 cm3 resolution can be obtained in 27 min [25, 32], and to retain the lowest SNR of 1.5 T at 3 T, the acquisition time can be shortened 26%, from 27 min to 17 min [25]. Using a high-sensitivity PA head coil, 3D MRSI with a voxel size of 0.34 cm3 can be acquired in 9.5 min [31]. Considering that gray matter is typically about 5 mm thick, a spatial resolution of 0.34 cm3 can allow the detection of differences between gray and white matter metabolite levels, avoiding partial volume effects. With a surface PA coil, 3D MRSI data with a spatial resolution as fine as 0.09 cm3 can be acquired in 17 min [31].
Example of 2D-multivoxel proton spectroscopic imaging (1H-MRSI) performed using a 3.0-T MR scanner (Signa Horizon LX) with a standard head coil, and the protocol (PROBE/SI) and the software (Functool Performance) provided by the manufacturer (General Electric Medical Systems, Milwaukee, WI). In brief, a point-resolved spectroscopy (PRESS) pulse sequence, with repetition time (TR) 1,500 ms, echo time (TE) 144 ms, field of view (FOV) 24 cm, phase 16, section thickness 10 mm and acquisition time 6 min 53 s, provided a spectroscopic data set, which was spatially zerofilled to 32 × 32 images with a final voxel resolution of 7.5 × 7.5 × 10 mm (0.56 cm3). Fluid-attenuated inversion recovery (FLAIR) (a) and contrast-enhanced T1-weighted (b) images from a 45-year-old man with a left fronto-temporo-parietal glioblastoma. Choline (Cho) (c), N-acetylaspartate (NAA) (d), creatine (Cr) (e) and lactate/lipids (LL) (f) maps. Localizing FLAIR image (g) and spectra (1–6) from the corresponding boxes in g. Color scale: red strongest signal intensity; blue weakest signal intensity. Higher spatial resolution improves the ability to recognize the spatial heterogeneity of high-grade gliomas. The spectrum 1, showing a low level of Cho, undetectability of Cr and NAA, and high LL peak, suggests the presence of necrosis. The spectrum 2, in the margins of glioma, presents the typical tumor pattern, characterized by an abnormal Cho/NAA ratio (<1). The spectra 3 and 4, in an apparently edematous region surrounding the enhanced margins of glioma, show low metabolite levels, as compared to the normal spectra 5 and 6, but different pattern: the spectrum 3 has a tumor pattern and suggests the presence of infiltrating tumor cells; the spectrum 4 has a normal Cho/NAA ratio and suggests noninfiltrated vasogenic edema
Spectral resolution and metabolite quantification
Spectral resolution, i.e., the ability to distinguish adjacent peaks in a spectrum, mainly depends on spectral distance, i.e., the difference in resonance frequency between two metabolites, and on attainable linewidth, i.e., the width at half-height of the metabolite resonance in Hz. The spectral distance (in Hz) is linearly correlated with the field strength and is about twice at 3 T as compared to 1.5 T (Fig. 2). The attainable linewidths are an inverse function of field-dependent T2 relaxation times and magnetic field homogeneity. Despite shorter T2 times and higher field inhomogeneity producing linewidths larger than at 1.5 T, the doubling of spectral distance at 3 T yields a better spectral resolution [25, 26]. This is reflected by improved baseline separation of Cho and Cr, which are only 0.2 parts per million (ppm) apart [25], and by better resolution of the glutamate/glutamine region, between 2.05 and 2.5 ppm, at shorter TE (Fig. 3) [26]. Nevertheless, despite the better spectral resolution and higher SNR, the reproducibility of metabolite measurements at 3 T is similar to [28, 33] or even worse than [27] that at 1.5 T. This shortfall can be explained by the larger linewidths at 3 T, decreasing both the SNR and the precision of quantitative analysis [27]. High spatial resolution at 3 T, however, reduces the magnetic field inhomogeneity and hampers the contraction of T2, decreasing the linewidths, especially for voxels below 0.75 cm3. This produces standard errors of the mean comparable in different measurements, suggesting the possibility of achieving high-precision quantification at those resolutions [32]. Unfortunately, studies focusing on the accuracy and precision of metabolite measurements at 3 T in high-resolution voxels are lacking.
Comparison of single-voxel spectra obtained with (c) a 1.5-T MR scanner (Signa Echo Speed) and (d) a 3-T scanner (Signa Horizon LX), using the protocol (PROBE-P) and the software provided by the manufacturer (General Electric Medical Systems, Milwaukee, WI). a Localizing T2-weighted image showing a low-grade glioma in the left temporal lobe of a 43-year-old woman. b Spectra recorded at 1.5 T (red) and 3 T (black), using PRESS sequence with TR 2,000 ms, TE 144 ms, 128 averages, voxel size 2.0 × 2.0 × 2.0 cm and acquisition time 4 min 56 s, and overlapped for emphasizing the differences. At 3 T, the signal-to-noise ratio (SNR) is about 25% higher and the spectral distance between the metabolites (in hertz) is doubled
Spectral resolution of the glutamate/glutamine (Glx) region with different proton spectroscopic approaches. a,b Localized spectra recorded in in vivo at 1.5 T and 3 T (scanners as in Fig. 2), using PRESS with TR 2,000 ms, TE 35 ms and 128 averages. The spectrum at 3 T shows higher SNR and better resolution of Glx region, between 2.1 and 2.5 parts per million (ppm). c,d,e TE-averaged PRESS: spectra form a phantom acquired by JPRESS with 16 different TE ranging from 35 to 195 ms (c), spreading the signal information into two spectral dimensions, and resulting spectrum (d); spectrum obtained in vivo from a healthy subject (e). TE-averaged PRESS fully resolves the glutamate (Glu) at 2.38 ppm from glutamine overlap, resulting in its unobstructed detection
The measurement of metabolite relaxation times is crucial for a reliable and reproducible determination of absolute metabolite concentration. While the T1 relaxation time of water protons is significantly longer at 3 T as compared to 1.5 T, metabolite T1 measurements can be either longer [34] or almost unchanged [35, 36]. The discrepancies can be explained by the large interindividual variability at 1.5 T and the possible differences in scanner performance, sequence design and strategies for acquiring and evaluating spectra. Another factor that can contribute to the variability is the T1 regional differences reported both at 3 T and 1.5 T [34, 36]. Ethofer et al. [34], for example, reported metabolite T1 values only slightly longer at 3 T vs. 1.5 T in the occipital lobes and cerebellum, and significantly increased at 3 T in the motor cortex, frontoparietal WM and thalamus (Table 1). Contrarily, metabolite T2 relaxation times at 3 T are markedly reduced as compared to 1.5 T (Table 1) and show distinct regional variations, especially for NAA. These region-specific variations and the different T2 effect on NAA, Cho and Cr have to be taken into account when using T2 values for absolute quantification. Furthermore, due to the decreased T2, MRS protocols with shorter TE values are preferable at 3 T, while TR adjustments appear of minor importance because of controversial T1 relaxation changes [36].
Static (B0) and radiofrequency (B1) magnetic field inhomogeneity
A wide variety of factors can alter the homogeneity of static (B0) and radiofrequency (B1) magnetic fields, affecting the quantitative and qualitative analyses of spectra. B0 inhomogeneities depend mainly on the effects linked to magnetic susceptibility, i.e., the ability of a substance to become magnetized. All paramagnetic and diamagnetic tissue compartments as well as implanted metallic structures affect B0 in different ways, inducing magnetic field distortions directly correlated to the applied magnetic field. Examples of susceptibility artifacts are those occurring near the air-filled sinuses, the skull base and the calvaria, provoked by susceptibility differences at tissue interfaces. A variety of susceptibility artifact correction methods have been proposed, including gradient compensation or z-shim techniques, tailored RF pulses, active and passive shimming and post-processing [42].
B1 inhomogeneities derive from several factors, including: imperfections in the RF pulse profile, an inhomogeneous transmit field, nonuniform reception sensitivity, RF penetration effects and eddy currents. These problems become more pronounced at high magnetic fields where RF wavelengths in tissue become shorter. Methods to reduce the effects of B1 inhomogeneity include: postprocessing with or without prior knowledge of the B1 field distribution; the use of tailored, adiabatic and composite RF pulses; special coil designs; the use of dielectric padding; optimizing current magnitudes and phases (RF shimming). These methods have both advantages and disadvantages [43, 44].
Eddy currents are small electric currents that are more prominent at high field strengths because of the increased speed and power of gradient coils. They can create an additional magnetic field and therefore cause spectral distortion and loss in SNR [45]. Furthermore, they can induce considerable torque on metallic instruments and implants, which can exceed the current limits of MR safety [46]. Besides dedicated hardware designs including magnetic field gradient and shim pulse-shaping networks [47], the effects of eddy currents can be minimized by correcting algorithms using the water signal as a reference [45]. Schulte et al. [48] have proposed an efficient 2D eddy current correction procedure, based on a 1D phase deconvolution method.
Chemical shift misregistration and J-modulation artifacts
Localization techniques, such as point-resolved spectroscopy (PRESS) or, to a lesser extent, stimulated-echo acquisition mode (STEAM), cause chemical shift displacement artifacts, which can give rise to signal misregistration for almost all metabolites. Only the signal from one specific frequency, usually NAA, originates from the selected VOI, while signals from molecules with different chemical shifts stem from spatially shifted volumes. Chemical shift displacement approximately scales with the square of the magnetic field strength, and this is partly due to reduced RF pulse bandwidths as well as increased chemical shift frequency separation [49]. There are several ways to minimize these artifacts, including the choice of frequency-encoding direction, field of view and receiver bandwith, and the use of fat-suppression techniques, such as inversion recovery sequences, very selective suppression (VSS) pulses, echo-planar spectroscopic imaging (EPSI), spectral spatial pulses and postprocessing approaches [49, 50]. Ozturk-Isik et al. [50] have recently proposed a postprocessing method, using a sensitivity-encoding (SENSE) algorithm, to reduce contamination from aliasing lipid resonances in 3D MRSI. This method is effective for data acquired at 3 T using a multichannel RF coil and high bandwidth, very selective saturation (VSS) pulses.
J-modulation anomalies refer to signal loss or cancellation of homonuclear-coupled resonances, arising from their chemical shift separation. The extent of the signal loss can vary considerably depending on the field strength, the used coil and the sequence parameters. One example is the doublet at 1.33 ppm of lactate, which can completely disappear at 3 T for TE 144 ms. Strategies to prevent or alleviate the signal loss due to anomalous J-modulation present both advantages and disadvantages [12, 49, 51].
Magnetic field stability
Magnetic field instability is another factor that can affect the quality of acquired spectra. In fact, even small drift of B 0 can result in measurable resonance frequency shift during the 1H-MRS acquisition, causing suboptimal water suppression, broadening of spectral lines and loss of phase coherence. Spectral distortion due to field instabilities, particularly concerning the high-field systems, can be compensated for by dedicated correction algorithms [52].
An additional source of instability at higher fields is the resonance frequency drifts induced by gradient heating. These drifts can be corrected by the method recently proposed by Ebel and Maudsley [53], which allows for obtaining both a nonlocalized B 0 measurement and a fully phase-encoded water reference signal, and then for correcting the frequency drift during postprocessing using the measured values. The correction largely improves spectral quality, removing lineshape distortions and recovering metabolite signal loss.
Physiological motion, such as breathing and cardiac/arterial pulsation, is another source of SNR loss and adverse consequences on 1H-MRS acquisition. However, its effects on brain PRESS at 3 T are negligible, implying that cardiac gating and breath pacing/gating are not routinely required for such an approach [54].
Safety issues
As B0 increases, the amount of RF power required by pulse sequences also increases and can entail several biological hazards. The specific absorption rate (SAR), namely the measure of energy absorbed from the effects of RF pulses, is four times higher at 3 T vs. 1.5 T and places some limits on the clinical setting. These can manifest as a reduction in slice number per TR period, longer scanner time and “patient cooling” delays between sequences. However, later-generation 3-T devices, which have new, more SAR-efficient system designs, innovative pulse sequence manipulations, parallel imaging and more appropriate imaging protocols, have limited the rate of RF energy deposition and should shortly lead to RF limitation and section acquisition equal to or slightly greater than those currently in place at 1.5 T [20]. A number of implants and devices (clips, stents, prostheses, etc.) have been assessed for magnetic field interactions at 3 T, and of these only 4% have been potentially unsafe [55].
Advanced 1H-MRS techniques at 3 T
Spectral editing
Any technique that simplifies the appearance of a spectrum for better observation and quantification of one or more metabolites of interest can be considered a spectral editing technique. Most of these techniques rely on the phenomenon of homonuclear or heteronuclear spin coupling, also known as J-coupling, which is responsible for several spectral patterns such as the doublet of lactate or the multiplet of γ-aminobutyric acid (GABA). Several editing sequences cannot be applied with 1.5-T scanners since they need higher SNR and sufficient spectral resolution. Examples of unresolved metabolites due to the J-coupling phenomenon are the Glx-components glutamate, glutamine and GABA, which cannot be easily differentiated from each other at 1.5 T and therefore have been frequently treated as a metabolite group [49, 51, 56–58].
Glutamate (Glu), the major excitatory neurotransmitter of the central nervous system, has neurotoxic properties, which have been associated with several neurodegenerative diseases. Glu gives rise to a complex spectrum characterized by the coupled spins of the C2–C4 hydrogen nuclei and the overlaps with other resonances, especially glutamine (Gln), glutathione and NAA. Glu and Gln, in particular, have a very similar chemical structure and overlap in their multiplet resonance groups. The editing approaches implemented at 3 T to resolve Glu include the multiple-quantum filter (MDF) procedure [56], more recently modified by Schubert et al. [57], and 2D J-resolved spectroscopy, dubbed JPRESS since it employs PRESS for volume localization [48]. The MQF method utilizes coherence transfer between different spin quantum states as well as gradient filtering to select the signal of one metabolite specifically and to suppress interfering signals. However, it has the disadvantage of simultaneously suppressing signals from Gln, NAA and myo-inositol, which are of diagnostic value [59]. JPRESS consists of a series of spin-echo experiments with different echo times spreading the signal information into two spectral dimensions [48]. Interesting variants of JPRESS implemented at 3 T include: chemical shift-selective filter (CSSF) [58], TE-averaged PRESS (Fig. 3) [59], constant time (CT) PRESS [60] and maximum-echo sampling [48]. All these procedures have been applied to a single voxel with a size ranging from 8 cm3 to 15.6 cm3. Srinivasan et al. [61] have recently proposed a TE-averaged PRESS technique for 2D 1H-MRSI (TE-averaged MRSI) to generate glu maps with a spatial resolution of 1.8 cm3 and a scan time of about 21 min.
Prior-knowledge fitting (ProFit) of J-resolved spectra is a novel method that utilizes a 2D fitting procedure, namely LCModel (linear combination of model spectra) in the frequency domain and VARPRO (variable projection) in the time domain, applied to JPRESS. Beside glutamate, it shows the detectability of a wide range of metabolites, including myo-inositol, glutathione, scyllo-inositol, GABA, alanine and ascorbic ac [62].
For selectively measuring both Glu and Gln, Choi et al. [63] have recently proposed a single-voxel, spectrally-selective refocusing method, which allowed the measurement of these metabolites with negligible contamination from NAA and glutathione.
GABA is the major inhibitor neurotransmitter in the human brain and has been implicated in several neurological and psychiatric disorders. Due to its low concentration, intense overlapping signals and complex spin system, GABA measurement is particularly challenging at field strengths accessible to clinical studies. A number of spectral editing techniques have been proposed in an attempt to resolve GABA. The techniques successfully implemented at 3 T include: single quantum difference spectroscopy (SQDS) [64], STEAM with optimized sequence times [65], maximum-echo sampling [48], double-quantum filtering with selective J rewinding [66] and ProFit of J-resolved spectra [62]. The selective homonuclear Hartmann-Hahn transfer method, recently proposed by Choi et al. [67], provides a spectral selectivity and a sensitivity greater than those reported by previous GABA-editing methods.
Diffusion tensor 1H-MRSI
In vivo diffusion spectroscopy can give insights into the physico-chemical properties of the intracellular environment. One example is the hypothesis that the water diffusion drop, associated with the development of ischemic cytotoxic edema, is caused not only by a water shift from extra-cellular to intra-cellular space, but also by reduced diffusion of intracellular water. This hypothesis arises from diffusion spectroscopic studies, which have demonstrated that purely intra-cellular metabolites, such as NAA, also show a reduction of diffusion without any compartmental shifts [68, 69]. Since diffusion for either water or metabolites is anisotropic in white matter, it can be characterized properly only by measuring the full diffusion tensor. However, such a measure implies lengthy acquisition times, which can be reduced only by acquiring spectra at a higher field. Recently, Ellegood et al. [70] measured the diffusion tensor of NAA, Cho and Cr at 3 T in six directions, in the brain of healthy volunteers, and demonstrated that the principal diffusion direction for NAA is consistent with the expected anatomic tract directions in the white matter.
Fast 1H-MRSI
For the purpose of improving the temporal resolution of metabolic brain mapping, different MR techniques, initially developed for fast conventional MRI, have been applied to multidimensional MRSI. Fast 1H-MRSI techniques implemented at 3 T include echoplanar spectroscopic imaging (EPSI) [71], parallel imaging with multicoil, multireceiver arrangements [50] and spiral acquisitions [72]. Each method comes with its own set of limitations and advantages and is under development in order to insure gainful application to the clinical routine. EPSI may be useful for identifying mechanisms underlying the blood oxygenation level dependence (BOLD) of the water signal during brain activation studies, but mapping the signal changes from brain metabolites is considerably more challenging [71]. Parallel acquisition techniques require a very demanding post-processing, and the resulting spectra are prone to artifacts or quality losses [73, 74]. In effect, although these techniques enable a significant reduction in acquisition time, the resulting spectra suffer from reduced SNR, and the need of multiple averaging to regain SNR prevents its application to the clinical setting. Figure 4 illustrates the results of a new promising technique using a prototype 3D PRESS sequence with an array spatial sensitivity encoding technique (ASSET). Mayer et al. [72] have recently presented a new promising method combining CT-PRESS with fast spiral MRSI. This method allows the acquisition of multivoxel spectra with a nominal in-plane resolution of 1.5 × 1.5 cm2 within 14 min, detecting signals from NAA, Cr, Cho, Glu and myo-inositol with no or only minor baseline distortions.
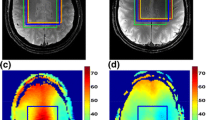
Demonstration of parallel acquisition technique using a prototype 3D PRESS sequence with an array spatial sensitivity encoding technique (ASSET). The experiments were performed on a 3.0-T MR scanner (GE EXCITE III HDTM 3 T) using an eight-channel receive-only head coil. The sequence performance was evaluated using a phantom containing water and lipid. The total acquisition time of the fully sampled data was 8 min 32 s (3D PRESS, TR = 1,000 ms, TE = 35 ms, FOV = 26 cm, 3D phase encoding steps =8 × 8 × 8) and could be reduced to 1 min 4 s by means of data undersampling with a reduction factor of two in all three spatial dimensions. All data were reconstructed and visualized using Matlab (The Mathworks, Version 7.1.0.124 SP 3). a Axial view of the phantom with the selected volume of interest. b Spectra from the selected region of interest. The dotted box shows the corresponding folded region of interest. c Spectrum within the green box showing original and undersampled data. Typically for the folding effect is the resulting summation of metabolite intensities, yielding higher peak amplitudes. d Reconstructed spectrum after 3D MRSI sensitivity encoding, which is in good agreement to the reference (fully sampled) spectrum
Conclusions
The advent of clinical scanners operating at 3 T is taking structural and functional imaging to new levels and is reinvigorating clinical brain spectroscopy. Many of the challenges that initially limited the diffusion of 3-T MR devices have been addressed by several strategies, the awareness of which is important to improve the efficiency of 3-T 1H-MRS and facilitate its application to the clinical routine. In particular, the development of multichannel-head coils that enables parallel imaging, the choice of proper field-specific sequence parameters, the refinement of correction algorithms and the application of special spectral editing approaches have greatly improved and added value to the diagnostic and research potentiality of MRS. However, continued software and hardware development and optimized sequence pulses are still needed to reach the maximal efficiency of 3-T brain 1H-MRS.
References
Bonavita S, Di Salle F, Tedeschi G (1999) Proton MRS in neurological disorders. Eur J Radiol 30:125–131
Del Sole A, Gambini A, Falini A, Lecchi M, Lucignani G (2002) In vivo neurochemistry with emission tomography and magnetic resonance spectroscopy: clinical applications. Eur Radiol 12:2582–2599
Lin A, Ross BD, Harris K, Wong W (2005) Efficacy of proton magnetic resonance spectroscopy in neurological diagnosis and neurotherapeutic decision making. NeuroRx 2:197–214
Sijens PE, Oudkerk M, Reijngoud DJ, Leenders KL, de Valk HW, van Spronsen FJ (2004) 1H MR chemical shift imaging detection of phenylalanine in patients suffering from phenylketonuria (PKU) Eur Radiol 14:1895–1900
Sijens PE, Verbruggen KT, Meiners LC, Soorani-Lunsing RJ, Rake JP, Oudkerk M (2005) 1H chemical shift imaging of the brain in guanidino methyltransferase deficiency, a creatine deficiency syndrome; guanidinoacetate accumulation in the gray matter. Eur Radiol 15:1923–1926
Huisman TA, Thiel T, Steinmann B, Zeilinger G, Martin E (2002) Proton magnetic resonance spectroscopy of the brain of a neonate with nonketotic hyperglycinemia: in vivo-in vitro (ex vivo) correlation. Eur Radiol 12:858–861
Bianchi MC, Tosetti M, Fornai F, Alessandri MG, Cipriani P, De Vito G, Canapicchi R (2000) Reversible brain creatine deficiency in two sisters with normal blood creatine level. Ann Neurol 47:511–513
Cordery RJ, Macmanus D, Godbolt A, Rossor MN, Waldman AD (2006) Short TE quantitative proton magnetic resonance spectroscopy in variant Creutzfeldt-Jakob disease. Eur Radiol 12:1–7
Hájek M, Adamovičová M, Herynek V, Škoch A, Jírů F, Křepelová A, Dezortová M (2005) MR relaxometry and 1H MR spectroscopy for the determination of iron and metabolite concentrations in PKAN patients. Eur Radiol 15:1060–1068
Sener RN (2003) Diffusion MRI and spectroscopy in Rasmussen’s encephalitis. Eur Radiol 13:2186–2191
Frayne R, Goodyear BG, Dickhoff P, Lauzon ML, Sevick RJ (2003) Magnetic resonance imaging at 3.0 Tesla: challenges and advantages in clinical neurological imaging. Invest Radiol 38:385–402
Hetherington HP, Pan JW, Chu W-J, Mason GF, Newcomer BR (1997) Biological and clinical MRS at ultra-high field. NMR Biomed 10:360–371
Di Costanzo A, Trojsi F, Tosetti M, Giannatempo GM, Nemore F, Piccirillo M, Bonavita S, Tedeschi G, Scarabino T (2003) High-field proton MRS of human brain. Eur J Radiol 48:146–153
Schick F (2005) Whole-body MRI at high field: technical limits and clinical potential. Eur Radiol 15:946–959
Schmitz BL, Aschoff AJ, Hoffmann MHK, Grön G (2005) Advantages and pitfalls in 3 T MR brain imaging: a pictorial review. AJNR Am J Neuroradiol 26:2229–2237
Ross JS (2004) The high-field-strength curmudgeon. AJNR Am J Neuroradiol 25:168–169
Pattany PM (2004) 3 T MR imaging: the pros and cons. AJNR Am J Neuroradiol 25:1455–1456
Shapiro MD, Magee T, Williams D, Ramnath R, Ross JS (2004) The time for 3 T clinical imaging is now. AJNR Am J Neuroradiol 25:1628–1629
DeLano MC, Fisher C (2006) 3 T MR imaging of the brain. Magn Reson Imaging Clin N Am 14:77–88
Tanenbaum LN (2006) Clinical 3 T MR imaging: mastering the challenges. Magn Reson Imaging Clin N Am 14:1–15
Schwindt W, Kugel H, Bachmann R, Kloska S, Allkemper T, Maintz D, Pfleiderer B, Tombach B, Heindel W (2003) Magnetic resonance imaging protocols for examination of the neurocranium at 3 T. Eur Radiol 13:2170–2179
Lu H, Nagae-Poetscher LM, Golay X, Lin D, Pomper M, van Zijl PCM (2005) Routine clinical brain MRI sequences for use at 3.0 Tesla. J Magn Reson Imaging 22:13–22
Trattnig S, Pinker K, Ba-Ssalamah A, Nöbauer-Huhmann IM (2006) The optimal use of contrast agents at high field MRI. Eur Radiol (Epub ahead of print)
Bachmann R, Reilmann R, Schwindt W, Kugel H, Heindel W, Krämer S (2006) FLAIR imaging for multiple sclerosis: a comparative MR study at 1.5 and 3.0 Tesla. Eur Radiol 16:915–921
Gonen O, Gruber S, Li BSY, Mlynárik V, Moser E (2001) Multivoxel 3D proton spectroscopy in the brain at 1.5 versus 3.0 T: signal-to-noise ratio and resolution comparison. Am J Neroradiol 22:1727–1731
Barker PB, Hearshen DO, Boska MD (2001) Single-voxel proton MRS of the human brain at 1.5 T and 3.0 T. Magn Reson Med 45:765–769
Kantarci K, Reynolds G, Petersen RC, Boeve BF, Knopman DS, Edland SD, Smith GE, Ivnik RJ, Tangalos EG, Jack CR Jr (2003) Proton MR spectroscopy in mild cognitive impairment and Alzheimer disease: comparison of 1.5 and 3 T. AJNR Am J Neuroradiol 24:843–849
Inglese M, Spindler M, Babb JS, Sunenshine P, Law M, Gonen O (2006) Field, coil, and echo-time influence on sensitivity and reproducibility of brain proton MR spectroscopy. AJNR Am J Neuroradiol 27:684–688
de Zwart JA, Ledden PJ, van Gelderen P, Bodurka J, Chu R, Duyn JH (2004) Signal-to-noise ratio and parallel imaging performance of a 16-channel receive-only brain coil array at 3.0 Tesla. Magn Reson Med 51:22–26
Sandgren N, Stoica P, Frigo FJ, Selen Y (2005) Spectral analysis of multichannel MRS data. J Magn Reson 175:79–91
Xu D, Chen AP, Cunningham C, Osorio JA, Nelson SJ, Vigneron DB (2006) Spectroscopic imaging of the brain with phased-array coils at 3.0 T. Magn Reson Imaging 24:69–74
Gruber S, Mlynárik V, Moser E (2003) High-resolution 3D proton spectroscopic imaging of the human brain at 3 T: SNR issues and application for anatomy-matched voxel sizes. Magn Reson Med 49:299–306
Wellard RM, Briellmann RS, Jennings C, Jackson GD (2005) Physiologic variability of single-voxel proton MR spectroscopic measurements at 3 T. AJNR Am J Neuroradiol 26:585–590
Ethofer T, Mader I, Seeger U, Helms G, Erb M, Grodd W, Ludolph A, Klose U (2003) Comparison of longitudinal metabolite relaxation times in different regions of the human brain at 1.5 and 3 Tesla. Magn Reson Med 50:1296–1301
Mlynárik V, Gruber S, Moser E (2001) Proton T 1 and T 2 relaxation times of human brain metabolites at 3 Tesla. NMR Biomed 14:325–331
Träber F, Block W, Lamerichs R, Gieseke J, Schild HH (2004) 1H metabolite relaxation times at 3.0 Tesla: measurements of T1 and T2 values in normal brain and determination of regional differences in transverse relaxation. J Magn Reson Imaging 19:537–545
Kreis R, Ernst T, Ross BD (1993) Development of the human brain: in vivo quantification of metabolite and water content with proton magnetic resonance spectroscopy. Magn Reson Med 30:424–437
Brief EE, Whittall KP, Li DKB, MacKay AL (2000) Metabolite T1 differs within and between regions of normal human brain. In: Proceedings of the 8th Annual Meeting of ISMRM, Denver, p 1939
Block W, Karitzky J, Träber F, Pohl C, Keller E, Mundegar RR, Lamerichs R, Rink H, Ries F, Schild HH, Jerusalem F (1998) Proton magnetic resonance spectroscopy of the primary motor cortex in patients with motor neuron disease: subgroup analysis and follow-up measurements. Arch Neurol 55:931–936
Frahm J, Bruhn H, Gyngell ML, Merboldt KD, Hänicke W, Sauter R (1989) Localized proton NMR spectroscopy in different regions of the human brain in vivo. Relaxation times and concentrations of cerebral metabolites. Magn Reson Med 11:47–63
Manton DJ, Lowry M, Blackband SJ, Horsman A (1995) Determination of proton metabolite concentrations and relaxation parameters in normal human brain and intracranial tumours. NMR Biomed 8:104–112
Truong T-K, Chakeres DW, Beversdorf DQ, Scharre DW, Schmalbrock P (2006) Effects of static and radiofrequency magnetic field inhomogeneity in ultra-high field magnetic resonance imaging. Magn Reson Imaging 24:103–112
Wang J, Qiu M, Yang QX, Smith MB, Constable RT (2005) Measurement and correction of transmitter and receiver induced nonuniformities in vivo. Magn Reson Med 53:408–417
Saekho S, Yip C-y, Noll DC, Boada FE, Stenger VA (2006) Fast-kz three-dimensional tailored radiofrequency pulse for reduced B1 inhomogeneity. Magn Reson Med 55:719–724
Simonetti AW, Melssen WJ, van der Graaf M, Heerschap A, Buydensa LMC (2002) Automated correction of unwanted phase jumps in reference signals which corrupt MRSI spectra after eddy current correction. J Magn Reson 159:151–157
Graf H, Lauer UA, Schick F (2006) Eddy-current induction in extended metallic parts as a source of considerable torsional moment. J Magn Reson Imaging 23:585–590
Gach HM, Lowe IJ, Madio DP, Caprihan A, Altobelli SA, Kuethe DO, Fukushima E (1998) A programmable pre-emphasis system. Magn Reson Med 40:427–431
Schulte RF, Lange T, Beck J, Meier D, Boesinger P (2006) Improved two-dimensional J-resolved spectroscopy. NMR Biomed 19:264–270
Lange T, Dydak U, Roberts TPL, Rowley HA, Bjeljac M, Boesiger P (2006) Pitfalls in lactate measurements at 3 T. AJNR Am J Neuroradiol 27:895–901
Ozturk-Isik E, Crane JC, Cha S, Chang SM, Berger MS, Nelson SJ (2006) Unaliasing lipid contamination for MR spectroscopic imaging of gliomas at 3 T using sensitivity encoding (SENSE). Magn Reson Med 55:1164–1169
Thrippleton MJ, Edden RAE, Keeler J (2005) Suppression of strong coupling artefacts in J-spectra. J Magn Reson 174:97–109
Thiel T, Czisch M, Elbel GK, Hennig J (2002) Phase coherent averaging in magnetic resonance spectroscopy using interleaved navigator scans: compensation of motion artifacts and magnetic field instabilities. Magn Reson Med 47:1077–1082
Ebel A, Maudsley AA (2005) Detection and correction of frequency instabilities for volumetric 1H echo-planar spectroscopic imaging. Magn Reson Med 53:465–469
Katz-Brull R, Lenkinski RE (2004) Frame-by-frame PRESS 1H-MRS of the brain at 3 T: the effects of physiological motion. Magn Reson Med 51:184–187
Shellock FG (2002) Biomedical implants and devices: assessment of magnetic field interactions with a 3.0-Tesla MR system. J Magn Reson Imaging 16:721–732
Thompson RB, Allen PS (1998) A new multiple quantum filter design procedure for use on strongly coupled spin systems found in vivo: its application to glutamate. Magn Reson Med 39:762–771
Schubert F, Gallinat J, Seifert F, Rinneberg H (2004) Glutamate concentrations in human brain using single voxel proton magnetic resonance spectroscopy at 3 Tesla. NeuroImage 21:1762–1771
Schulte RF, Trabesinger AH, Boesiger P (2005) Chemical-shift-selective filter for the in vivo detection of J-coupled metabolites at 3 T. Magn Reson Med 53:275–281
Hurd R, Sailasuta N, Srinivasan R, Vigneron DB, Pelletier D, Nelson SJ (2004) Measurement of brain glutamate using TE-averaged PRESS at 3 T. Magn Reson Med 51:435–440
Mayer D, Spielman DM (2005) Detection of glutamate in the human brain at 3 T using optimized constant time point resolved spectroscopy. Magn Reson Med 54:439–442
Srinivasan R, Cunningham C, Chen A, Vigneron D, Hurd R, Nelson SJ, Pelletier D (2006) TE-Averaged two-dimensional proton spectroscopic imaging of glutamate at 3 T. NeuroImage 30:1171–1178
Schulte RF, Boesiger P (2006) ProFit: two-dimensional prior-knowledge fitting of J-resolved spectra. NMR Biomed 19:255–263
Choi C, Coupland NJ, Bhardwaj PP, Malykhin N, Gheorghiu D, Allen PS (2006) Measurement of brain glutamate and glutamine by spectrally-selective refocusing at 3 Tesla. Magn Reson Med 55:997–1005
Henry P-G, Dautry C, Hantraye P, Bloch G (2001) Brain GABA editing without macromolecule contamination. Magn Reson Med 45:517–520
Hanstock CC, Coupland NJ, Allen PS (2002) GABA X2 multiplet measured pre- and post-administration of vigabatrin in human brain. Magn Reson Med 48:617–623
Choi C, Coupland NJ, Hanstock CC, Ogilvie CJ, Higgins ACM, Gheorghiu D, Allen PS (2005) Brain γ-aminobutyric acid measurement by proton double-quantum filtering with selective J rewinding. Magn Reson Med 54:272–279
Choi I-Y, Lee S-P, Shen J (2005) Selective homonuclear Hartmann-Hahn transfer method for in vivo spectral editing in the human brain. Magn Reson Med 53:503–510
Dijkhuizen RM, de Graaf RA, Tulleken KA, Nicolay K (1999) Changes in the diffusion of water and intracellular metabolites after excitotoxic injury and global ischemia in neonatal rat brain. J Cereb Blood Flow Metab 19:341–349
Dreher W, Busch E, Leibfritz D (2001) Changes in apparent diffusion coefficients of metabolites in rat brain after middle cerebral artery occlusion measured by proton magnetic resonance spectroscopy. Magn Reson Med 45:383–389
Ellegood J, Hanstock CC, Beaulieu C (2006) Diffusion tensor spectroscopy (DTS) of human brain. Magn Reson Med 55:1–8
Mulkern RV, Chen NK, Oshio K, Panych LP, Rybicki FJ, Gambarota G (2004) Fast spectroscopic imaging strategies for potential applications in fMRI. Magn Reson Imaging 22:1395–1405
Mayer D, Kim D-H, Adalsteinsson E, Spielman DM (2006) Fast CT-PRESS-based spiral chemical shift imaging at 3 Tesla. Magn Reson Med 55:974–978
Dydak U, Pruessmann KP, Weiger M, Tsao J, Meier D, Boesiger P (2003) Parallel spectroscopic imaging with spin-echo trains. Magn Reson Med 50:196–200
Sánchez-González J, Tsao J, Dydak U, Desco M, Boesiger P, Pruessmann KP (2006) Minimum-norm reconstruction for sensitivity-encoded magnetic resonance spectroscopic Imaging. Magn Reson Med 55:287–295
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 2.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by-nc/2.0/.
About this article
Cite this article
Di Costanzo, A., Trojsi, F., Tosetti, M. et al. Proton MR spectroscopy of the brain at 3 T: an update. Eur Radiol 17, 1651–1662 (2007). https://doi.org/10.1007/s00330-006-0546-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-006-0546-1