Abstract
To describe signal and contrast properties of an isotropic, single-slab 3D dataset [double inversion-recovery (DIR), fluid-attenuated inversion recovery (FLAIR), T2, and T1-weighted magnetization prepared rapid acquisition gradient-echo (MPRAGE)] and to evaluate its performance in detecting multiple sclerosis (MS) brain lesions compared to 2D T2-weighted spin-echo (T2SE). All single-slab 3D sequences and 2D-T2SE were acquired in 16 MS patients and 9 age-matched healthy controls. Lesions were scored independently by two raters and characterized anatomically. Two-tailed Bonferroni-corrected Student’s t-tests were used to detect differences in lesion detection between the various sequences per anatomical area after log-transformation. In general, signal and contrast properties of the 3D sequences enabled improved detection of MS brain lesions compared to 2D-T2SE. Specifically, 3D-DIR showed the highest detection of intracortical and mixed WM-GM lesions, whereas 3D-FLAIR showed the highest total number of WM lesions. Both 3D-DIR and 3D-FLAIR showed the highest number of infratentorial lesions. 3D-T2 and 3D-MPRAGE did not improve lesion detection compared to 2D-T2SE. Multi-contrast, isotropic, single-slab 3D MRI allowed an improved detection of both GM and WM lesions compared to 2D-T2SE. A selection of single-slab 3D contrasts, for example, 3D-FLAIR and 3D-DIR, could replace 2D sequences in the radiological practice.
Similar content being viewed by others
Introduction
Histopathological studies of patients with multiple sclerosis (MS) have provided evidence for lesions not only in the white matter (WM), but also in the cortical and deep gray matter (GM) [1–6]. Magnetic resonance imaging (MRI) plays a prominent role in detecting and monitoring MS lesions in the brain. Quantitative analyses based on two-dimensional (2D) T2-weighted spin-echo (T2SE) techniques (for example, T2 lesion loads and counts of active T2 lesions in longitudinal comparisons) have been investigated as possible surrogate markers for clinical disease progression. However, cross-sectional and longitudinal analyses, focusing on focal white matter pathology, have generally shown only mild to moderate correlations to clinical measures [7, 8]. The fact that 2D-T2SE is relatively insensitive for detecting MS lesions in some parts of the brain, particularly the cortical GM [9], combined with imperfect registration to compensate for repositioning errors, likely contributes to the poor clinico-radiological correlations found so far. Hence, new MRI techniques are needed that improve the detection of MS lesions in the brain and allow for a more accurate means of longitudinal comparison.
The detection of MS lesions may be improved in two ways: (1) by enhancing lesion-to-background contrast and (2) by improving spatial resolution. Two-dimensional (2D)-fluid-attenuated inversion-recovery (FLAIR) nulls the signal of cerebrospinal fluid (CSF), thereby increasing contrast between lesions and adjacent CSF. Studies using 2D-FLAIR indeed demonstrated an increased detection of WM lesions compared to 2D-T2SE, especially so for subcortical lesions, although at the cost of infratentorial lesion detection [10]. The 2D double inversion recovery (DIR) technique combines two inversion pulses that suppress the signals of both CSF and WM, creating an image with a clear cortical delineation and demonstrated high lesion conspicuity [11]. Regarding the improvement of spatial resolution, three-dimensional (3D) MRI allows for the acquisition of smaller (isotropic) voxels while maintaining a good signal-to-noise ratio (SNR). Initially, FLAIR and DIR could only be implemented as multi-slab 3D sequences; nevertheless, FLAIR demonstrated an improved detection of WM lesions [12, 13], and DIR showed an improved detection and classification of cortical lesions [14], all compared to 2D-T2SE. A recent paper showed that, aside from with DIR, cortical lesions could also be visualized employing a 3D T1-weighted sequence [15]. In spite of improved lesion detection, the relatively long acquisition times of multi-slab 3D sequences have so far hampered their introduction into the clinical setting. The advent of single-slab 3D methods, which use very long echo trains and refocusing pulses with variable flip angles, allowed whole-brain acquisition of FLAIR and T2 images [16]. The advantages of single-slab 3D imaging over multi-slab 3D imaging are a significant reduction of acquisition times and the absence of venetian-blind and flow artifacts. Recently, a single-slab 3D-DIR sequence was developed in order to expand the range of single-slab 3D-based contrast mechanisms for MS lesion detection [17]. So far, single-slab 3D sequences have not been comprehensively evaluated to determine their sensitivity in the detection of MS lesions in various parts of the brain. Furthermore, they have not been compared to a standard imaging technique such as 2D-T2SE, which is used routinely in both clinical treatment trials and the clinical radiological setting and has as such been incorporated into various MR imaging guidelines in MS [18–20].
Hence, the goal of the current study was to describe signal and contrast properties of an isotropic, single-slab 3D dataset (DIR, FLAIR, T2, and T1-MPRAGE) and to prospectively evaluate its performance in detecting MS brain lesions compared to a 2D-T2SE sequence.
Materials and methods
Patients and controls
Sixteen patients (9 women) with clinically definite MS [21], of which 5 with secondary progressive MS and 11 with relapsing-remitting MS, were randomly selected from a clinical database. Mean age of patients was 39.5 years (range 24.6–56.6), median expanded disability status scale score (EDSS) [22] was 2.0 (range 1.0–6.5), and mean disease duration was 10.4 years (range 4.0–26.4). In addition, nine age-matched healthy controls (three women), mean age of 32.0 years (range 22.9–53.5), were examined. The study protocol was approved by the local ethics review board, and all subjects gave written informed consent before investigation.
MR image acquisition
Imaging was performed on a 1.5 T whole body MR system (Siemens Sonata, Erlangen, Germany), using a standard circularly polarized head coil. Five sequences were acquired per subject in pseudo-randomized order, 2D dual-echo T2SE, single-slab 3D-DIR, 3D-FLAIR, 3D-T2 and a 3D-MPRAGE. The single-slab 3D sequences have very long echo trains, made possible by the use of refocusing pulses with variable flip angles [16]. As a result, the effective echo time of 349 ms produces a T2 contrast approximately equivalent to a TE of 100 ms in 2D fast spin-echo sequences [23]. Because all radio frequency pulses in the single-slab 3D sequences are non-spatially selective, the 3D slab was placed in the sagittal orientation, with read-out in the head-to-feet direction to prevent infolding. For more technical details and artifact descriptions refer to [16] and [17]. To obtain a high spatial resolution within clinically acceptable acquisition times (below 10 min), the inversion times for 3D-FLAIR and 3D-DIR were optimized at a chosen TR of 6500 ms. Sequence parameters and acquisition times are listed in Table 1.
Image analysis
The sagittal 3D images were orthogonally reformatted (i.e., without interpolation) into the axial plane. All images were analysed using a digital workstation (Centricity Radiology RA 600, GE Medical Systems, Fairfield, CT). First, images from all controls were analyzed to identify possible artifacts and abnormalities. Subsequently, a training session was held, in the presence of an experienced reader, to ensure that both independent readers used the same criteria to characterize GM and WM lesions within the different anatomical areas. Images were scored independently by the two readers, with the order of the sequences being randomized, to avoid recall bias. Lesions were characterized anatomically as follows: intracortical, mixed WM-GM, juxtacortical, deep GM (including the internal capsule), periventricular WM (directly abutting the ventricles or clearly connected to lesions abutting the ventricles), deep WM, and infratentorial. In addition, three combined areas were defined: total cortical (intracortical + mixed WM-GM + juxtacortical), total WM (periventricular WM + deep WM), and overall (total of all anatomical areas). Lesions extending over multiple slices were scored only once.
Signal and contrast properties
Signal intensities (SIs) were determined by placing regions of interest (ROIs) on the original datasets. ROIs were placed in lesions (only in WM lesions because GM lesions generally have the same signal intensities as WM lesions and are commonly too small for accurate ROI-based measures), WM, GM (hippocampus, amygdala and cingulate gyrus), CSF, and air (noise). Signal-to-noise ratios (SNRs) were calculated as: SNR = SI/Standard Deviation of the noise (SDnoise). The SDnoise was corrected for the underestimation that is due to magnitude reconstruction of the images [24]. In addition, lesion contrast ratios (CRs) were determined between lesion and WM and between lesion and GM using the following equation: CR = |SI1-SI2|/SI2, where SI1 denotes the signal intensity of lesion.
Statistical analysis
Per patient, the averages of the independent lesion counts of both readers, for each anatomical area and sequence, were calculated. Subsequently, these data were used to calculate the mean and standard deviation (SD) of lesion numbers of all patients per anatomical area and sequence. Because data were not normally distributed, a log transformation was applied. Two-tailed paired Student’s t-tests, adjusted for five pairwise comparisons (Bonferroni), were used to test for statistical differences among the five sequences per anatomical region. Corrected p-values of less than 0.05 were considered statistically significant. The intraclass correlation coefficient (ICC) [25] was calculated, using the log transformed data, with a variance component analysis to express interobserver agreement. The ICC ranges from zero to one with a higher ICC indicating a higher level of agreement. All statistical calculations were performed with SPSS version 12.0 (SPSS, Chicago, IL).
Results
Control subjects
Images from healthy controls, particularly the 3D-DIR and 3D-FLAIR images, contained several areas of intrinsically high signal intensity, such as the choroid plexus (Fig. 1). Small caps around the ventricles (Fig. 1) were also frequently seen, as described previously, for multi-slab 3D sequences [13, 14]. On the 3D-MPRAGE images vascular structures were recognizable as circular areas of low signal intensity in and around the cortex. The mean number for overall amount of lesions scored per control subject, presumably of vascular origin, was 0.3 lesions on 3D-DIR, 0.4 on 3D-FLAIR, 0.2 on 3D-T2, 0.1 on 3D-MPRAGE, and 0.2 on 2D-T2SE.
Two axially reformatted images from the same young healthy control, no contrast applied. A: 3D-FLAIR (TR/TE/TI 6500/349/2200 ms) and B: 3D-DIR (TR/TE/TI1/TI2 6500/349/2350/350 ms). Note two typical areas with high signal intensity on both sequences, arising from the choroid plexus (straight arrows) and from periventricular transependymal effusion (arrowheads)
MS patients
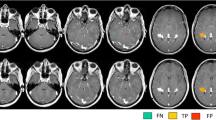
The mean number (SD) for overall amount of lesions scored per patient was 88.8 (67.8) on 3D-DIR, 100.4 (72.3) on 3D-FLAIR, 77.8 (54.6) on 3D-T2, 83.5 (71.9) on 3D-MPRAGE, and 82.4 (60.6) on 2D-T2SE images. The mean numbers per anatomical area, including statistical differences, are listed in Table 2. The highest numbers of intracortical and mixed WM-GM lesions were scored on 3D-DIR (Fig. 2). Both findings were significantly higher compared to 2D-T2SE (p = 0.036 and p = 0.019, respectively). By comparison, the lowest number of juxtacortical lesions was seen on 3D-DIR, whereas the most were scored on 2D-T2SE images; however, this difference was not significant (p = 0.086). The highest number of WM lesions was scored on 3D-FLAIR, which was significant compared to 2D-T2SE (p = 0.002). 3D-DIR showed the smallest number of deep WM lesions, although not significantly compared to 2D-T2SE. Both 3D-DIR and 3D-FLAIR, although not significant when compared to 2D-T2SE (p = 0.237 and p = 0.173, respectively), showed the highest detection of infratentorial lesions (Fig. 3).
Four axially reformatted images from an MS patient, no contrast applied. A: 3D-DIR, B: 3D-FLAIR, C: 3D-T2, and D: 3D-MPRAGE (sequence parameters are listed in Table 1). Images show several gray matter and white matter lesions. Straight arrows point out a juxtacortical lesion. Delta arrows point out a mixed WM-GM matter lesion that invades the cortical mantle. Arrowheads point out an intracortical lesion clearly seen on 3D-DIR and to a lesser extent on the other images
Five axial images from an MS patient, no contrast applied. A: 3D-DIR, B: 3D-FLAIR, C: 3D-T2, D: 3D-MPRAGE, and E: first echo 2D-T2SE (sequence parameters are listed in Table 1). The 2D-T2SE image had a slice thickness of 3 mm, whereas the 3D sequences had a 1.3 mm slice thickness. Note the flow artifacts in the 2D-T2SE image from the basilar artery (straight arrows) and from the left transverse sinus (delta arrow), and note the absence of flow artifacts in the 3D images. Also note the lesions in the pons and cerebellar peduncles and in the left trigeminal nerve (arrowheads)
Intraclass correlation coefficient
For the total cortical region the ICCs were highest for 3D-DIR (0.82) followed by 3D-MPRAGE (0.79). For the total WM region ICCs were highest for 3D-MPRAGE (0.86) and 3D-FLAIR (0.77). The ICCs for overall lesion counts were 3D-DIR (0.81), 3D-FLAIR (0.86), 3D-T2 (0.72), 3D-MPRAGE (0.88), and 2D-T2SE (0.64).
Signal and contrast properties
Mean SNRs and CRs, as well as their respective standard deviations, are listed in Table 3. The 3D-DIR showed the highest CR for Lesion-WM (10.8 ± 2.8), corresponding with the lowest SNR for WM (2.7 ± 1.0) (and CSF), due to successful signal suppression.
Discussion
Recent technological advances have made it possible to acquire a multi-contrast, isotropic, single-slab 3D dataset. In the current study we assessed this dataset for its signal and contrast properties and its benefit in the detection of different types of MS brain lesions compared to a 2D-T2SE sequence. 3D sequences, with their intrinsically higher SNR and small, nearly isotropic voxel dimensions, have several intrinsic advantages for post-processing purposes over 2D sequences. The 3D images can be easily reformatted without loss of image quality, which allows the selection of optimal viewing planes for various anatomical structures (see, e.g., Fig. 4). In addition, images can be registered more accurately, allowing for a more precise determination of spatiotemporal disease activity, which is important for longitudinal studies. Finally, the absence of flow artifacts improves lesion visualization in the posterior cranial fossa.
Twelve images from an MS patient, no contrast applied. A: 3D-DIR, B: 3D-FLAIR, C: 3D-T2, and D: 3D-MPRAGE (sequence parameters are listed in Table 1). First and second columns show axial and coronal reformatted MR images, respectively; third column shows original sagittal images. Due to the near isotropic resolution, image quality is preserved in all orientations. Note the (intra)cortical lesion in the right posterior cingulate gyrus (arrowheads)
The evaluation of this 3D dataset showed an increased detection of both gray and white matter lesions in comparison with a 2D-T2SE sequence. A conducive factor is the decrease in slice thickness, which is strongly associated with increased lesion detection [26, 27]. Several authors have previously reported increased detection of WM lesions using 3D multi-slab FLAIR sequences [12, 13, 26]. In our study, the highest number of WM lesions was also detected on 3D-FLAIR. Moreover, significantly more lesions were detected in the deep WM with 3D-FLAIR, compared to 3D-DIR. This was especially distinct for small lesions and may be explained by partial suppression of WM lesions (due to the combination of the two inversion pulses of the 3D-DIR sequence) and reduced intensity of partial volume effects around lesions. In this study the highest numbers of infratentorial lesions were scored on 3D-DIR and 3D-FLAIR images, which is in contrast with previously reported decreased detection in the posterior cranial fossa [10, 12, 13]. As stated above, this is related to the absence of blood and CSF flow artifacts in the 3D sequences, especially in the posterior cranial fossa (Fig. 3), which is explained by the single-slab nature of the sequence employing non-spatially selective radio-frequency pulses [28]. This is an important finding, as infratentorial lesions were found to strongly predict long-term disability in patients with initial findings suggestive of MS [29].
3D-DIR allowed an improved detection of lesions located intracortically or at the border of WM and GM (mixed WM-GM). The highest number of juxtacortical lesions was detected on 2D-T2SE, which likely represents a re-classification phenomenon. Lesions classified on 2D-T2SE images as juxtacortical were often seen to invade the cortical mantle on 3D-DIR images and were therefore scored as mixed WM-GM lesions. These results confirm the previously reported improved detection and classification of cortical MS lesions using multi-slab 3D-DIR [16]. Although the CR for lesion-GM was lower on the 3D-DIR (0.5 ± 0.1) compared to 3D-T2 (0.6 ± 0.2), the CR for lesion-CSF was much higher (10.6 ± 3.1) compared to 3D-T2 (0.2 ± 0.1), which indicates that the surrounding high signal of CSF on the 3D-T2 obscured (intra)cortical lesion detection. In accordance with [15], we observed both intracortical and mixed WM-GM lesions as hypointense areas on the T1-weighted 3D-MPRAGE. Interestingly, several cortical lesions were observed that were clearly visible on 3D-MPRAGE images, but were hardly or not visible on 3D-DIR and vice versa. Spatial variations in the T2 relaxation times throughout the cortical ribbon [30] might influence cortical lesion conspicuity and/or the 3D-MPRAGE might be more sensitive to lesions that represent a more chronic form of disease, in which prolongation of T1 relaxation time is more prominent than its T2 counterpart. Future studies will have to further investigate the nature of T1 hypointensity of cortical lesions and the benefit of the 3D-MPRAGE for the detection of black holes.
The images in this study were independently scored by two observers, and ICCs were calculated to express interobserver agreement. The overall highest ICC was calculated for 3D-MPRAGE (0.88) followed by 3D-FLAIR (0.86) and 3D-DIR (0.81), compared to an ICC of 0.64 for 2D-T2SE. Besides improved detection, the 3D sequences also appear very robust with ICCs indicating a more than good level of agreement between raters.
The shorter acquisition times of the single-slab method, for example, 9.8 min for the single-slab 3D-DIR versus 15.3 min for the multi-slab DIR [14], facilitate the introduction of this method into the clinical setting. Although 1.5 T is still the dominant clinical field strength, 3 T scanners are becoming more common. In a study comparing 3 T to 1.5 T, an increase of 13% in WM lesion detection was found for both 2D-FLAIR and T2SE [31]. The current study was performed at 1.5 T, using a standard circularly polarized head coil. Increased SNR might be obtained either by increasing the field strength or by using multi-channel phased array coils. Especially the 3D-DIR sequence will benefit from higher SNR, because of its intrinsically low SNR. For the 3D-FLAIR, 3D-T2 and 3D-MPRAGE, the increase in SNR can be used either to increase spatial resolution or to decrease acquisition time (by using parallel acquisition techniques). On the other hand, the decrease in acquisition times and slice thickness while maintaining or even improving spatial resolution and SNR will result in an increasing number of images that need to be analyzed. While advantageous for post-processing techniques and research aims, this development will put an increasing strain on the radiological practice and gives impetus to introduce and improve techniques that visualize only areas of change, such as image subtraction [32].
Besides reducing acquisition times, by use of, e.g., parallel imaging techniques, a choice can also be made for specific contrasts from the 3D dataset. In case of time constraints, for example, in an acute clinical setting, acquisition times can be reduced to less than 20 min by applying only 3D-FLAIR and 3D-DIR. These sequences provided the highest sensitivity for GM, WM, and infratentorial lesions, whereas 3D-T2 and 3D-MPRAGE did not demonstrate an improved detection of lesions in any of the anatomical areas compared to any of the other sequences, in particular 2D-T2SE. The reported increased sensitivity for lesions in various parts of the brain could lead to an earlier diagnosis of the disease as MRI criteria are strongly embedded in the diagnostic criteria for MS [33, 34]. Indeed, this was recently reported, although with the use of 2D sequences applied at higher field strength (3 T) instead of applying single-slab 3D imaging at 1.5 T [35]. Furthermore, the increased sensitivity for (intra)cortical lesions using 3D-DIR could improve the clinico-radiological correlations as lesions in eloquent gray matter areas could substantially contribute to clinical disability. Future studies with a larger sample size would have to evaluate the specific benefits of multi-contrast, single-slab 3D imaging with respect to clinical correlations and an earlier diagnosis of the disease. A future clinical protocol based on the single-slab 3D dataset presented here would ideally also include contrast-enhanced 3D-MPRAGE images to demonstrate disruption of the blood-brain barrier. Furthermore, such a protocol could include quantitative MR techniques such as magnetization transfer (MTR) and diffusion tensor imaging (DTI), and what the additional value of these techniques is for the identification of focal pathology compared to multi-contrast, single-slab 3D imaging should be examined.
In this paper, the multi-contrast nature of the protocol could have been exploited more fully, by one-on-one lesion evaluation of all four contrasts simultaneously. However, the primary goal of the current study was to describe general signal and contrast properties of the 3D dataset, as well as its overall performance in detecting lesions in different anatomical areas. Future studies will have to demonstrate the specific benefits of each separate contrast (or combinations thereof) for various post-processing purposes. Furthermore, it would have been possible to reslice the 3D sequences in the orientation of the 2D sequence to equate slice orientation and thickness. However, such a strategy would have degraded image quality and would not have optimally utilized the intrinsic ability of the single-slab 3D method to acquire thin slices with good SNR.
In conclusion, a multi-contrast, single-slab 3D dataset with near-isotropic voxels was presented and evaluated in MS patients. An increased detection of both GM and WM lesions was found compared to 2D-T2SE. 3D-DIR showed the highest detection of intracortical and mixed WM-GM lesions, while 3D-FLAIR allowed for the highest detection of WM lesions. Due to the absence of flow artifacts in the posterior cranial fossa, both 3D-DIR and 3D-FLAIR enabled an improved visualization of infratentorial lesions. Furthermore, (intra)cortical lesions were also visualized with a 3D T1-weighted sequence (MPRAGE). Finally, a selection of single-slab 3D contrasts, for example 3D-FLAIR and 3D-DIR, could replace 2D sequences in the radiological practice.
References
Kidd D, Barkhof F, McConnell R, Algra PR, Allen IV, Revesz T (1999) Cortical lesions in multiple sclerosis. Brain 122:17–26
Huitinga I, De Groot CJ, Van der Valk P, Kamphorst W, Tilders FJ, Swaab DF (2001) Hypothalamic lesions in multiple sclerosis. J Neuropathol Exp Neurol 60:1208–1218
Bø L, Vedeler CA, Nyland H, Trapp BD, Mørk SJ (2003) Intracortical multiple sclerosis lesions are not associated with increased lymphocyte infiltration. Mult Scler 9:323–331
Kutzelnigg A, Lucchinetti CF, Stadelmann C et al (2005) Cortical demyelination and diffuse white matter injury in multiple sclerosis. Brain 128:2705–2712
Kutzelnigg A, Lassmann H (2006) Cortical demyelination in multiple sclerosis: a substrate for cognitive deficits? J Neurol Sci 245:123–126
Wegner C, Esiri MM, Chance SA, Palace J, Matthews PM (2006) Neocortical neuronal, synaptic, and glial loss in multiple sclerosis. Neurology 67:960–967
Barkhof F (1999) MRI in multiple sclerosis: correlation with expanded disability status scale (EDSS). Mult Scler 5:283–286
Goodin DS (2006) Magnetic resonance imaging as a surrogate outcome measure of disability in multiple sclerosis: have we been overly harsh in our assessment? Ann Neurol 59:597–605
Geurts JJ, Bö L, Pouwels PJ et al (2005) Cortical lesions in multiple sclerosis: combined postmortem MR imaging and histopathology. AJNR Am J Neuroradiol 26:572–577
Filippi M, Yousry T, Baratti C et al (1996) Quantitative assessment of MRI lesion load in multiple sclerosis. A comparison of conventional spin-echo with fast fluid-attenuated inversion recovery. Brain 119:1349–1355
Bedell BJ, Narayana PA (1998) Implementation and evaluation of a new pulse sequence for rapid acquisition of double inversion recovery images for simultaneous suppression of white matter and CSF. J Magn Reson Imaging 8:544–547
Tubridy N, Barker GJ, Macmanus DG, Mosely IF, Miller DH (1998) Three-dimensional fast fluid attenuated inversion recovery (3D fast FLAIR): a new MRI sequence which increases the detectable lesion load in multiple sclerosis. Br J Radiol 71:840–845
Tan IL, Pouwels PJ, van Schijndel RA, Adèr HJ, Manoliu RA, Barkhof F (2002) Isotropic 3D fast FLAIR imaging of the brain in multiple sclerosis: initial experience. Eur Radiol 12:559–567
Geurts JJ, Pouwels PJ, Uitdehaag BM, Polman CH, Barkhof F, Castelijns JA (2005) Intracortical lesions in multiple sclerosis: improved detection with 3D double inversion-recovery MR imaging. Radiology 236:254–260
Bagnato F, Butman JA, Gupta S et al (2006) In vivo detection of cortical plaques by MR imaging in patients with multiple sclerosis. AJNR Am J Neuroradiol 27:2161–2167
Mugler JP 3rd, Bao S, Mulkern RV et al (2000) Optimized single-slab three-dimensional spin-echo MR imaging of the brain. Radiology 216:891–899
Pouwels PJ, Kuijer JP, Mugler JP 3rd, Guttmann CR, Barkhof F (2006) Human gray matter: feasibility of single-slab 3D double inversion-recovery high-spatial-resolution MR Imaging. Radiology 241:873–879
Filippi M, Horsfield MA, Adèr HJ et al (1998) Guidelines for using quantitative measures of brain magnetic resonance imaging abnormalities in monitoring the treatment of multiple sclerosis. Ann Neurol 43:499–506
Filippi M, Rocca MA, Arnold DL et al (2006) EFNS guidelines on the use of neuroimaging in the management of multiple sclerosis. Eur J Neurol 13:313–325
Simon JH, Li D, Traboulsee A et al (2006) Standardized MR imaging protocol for multiple sclerosis: Consortium of MS Centers consensus guidelines. AJNR Am J Neuroradiol 27:455–461
Poser CM, Paty DW, Scheinberg L et al (1983) New diagnostic criteria for multiple sclerosis: guidelines for research protocols. Ann Neurol 13:227–231
Kurtzke JF (1983) Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology 33:1444–1452
Mugler JP 3rd, Kiefer B, Brookeman JR (2000) Three-dimensional T2-weighted imaging of the brain using very long spin-echo trains [abstr]. In: Proceedings of the eighth meeting of the international society for magnetic resonance in medicine. Berkeley, Calif: International society for magnetic resonance in medicine, 687
Henkelman RM (1985) Measurement of signal intensities in the presence of noise in MR Images. Med Phys 12:232–233
Carrasco JL, Jover L (2003) Estimating the generalized concordance correlation coefficient through variance components. Biometrics 59:849–858
Molyneux PD, Tubridy N, Parker GJ et al (1998) The effect of section thickness on MR lesion detection and quantification in multiple sclerosis. AJNR Am J Neuroradiol 19:1715–1720
Dolezal O, Dwyer MG, Horakova D et al (2007) Detection of cortical lesions is dependent on choice of slice thickness in patients with multiple sclerosis. Int Rev Neurobiol 79:475–489
Kallmes DF, Hui FK, Mugler JP 3rd (2001) Suppression of cerebrospinal fluid and blood flow artifacts in FLAIR MR imaging with a single-slab three-dimensional pulse sequence: initial experience. Radiology 221:251–255
Minneboo A, Barkhof F, Polman CH, Uitdehaag BM, Knol DL, Castelijns JA (2004) Infratentorial lesions predict long-term disability in patients with initial findings suggestive of multiple sclerosis. Arch Neurol 61:217–221
Georgiades CS, Itoh R, Golay X, van Zijl PC, Melhem ER (2001) MR imaging of the human brain at 1.5 T: regional variations in transverse relaxation rates in the cerebral cortex. AJNR Am J Neuroradiol 22:1732–1737
Wattjes MP, Lutterbey GG, Harzheim M et al (2006) Higher sensitivity in the detection of inflammatory brain lesions in patients with clinically isolated syndromes suggestive of multiple sclerosis using high field MRI: an intraindividual comparison of 1.5 T with 3.0 T. Eur Radiol 16:2067–2073
Tan IL, van Schijndel RA, Pouwels PJ, Adèr HJ, Barkhof F (2002) Serial isotropic three-dimensional fast FLAIR imaging: using image registration and subtraction to reveal active multiple sclerosis lesions. AJR Am J Roentgenol 179:777–782
McDonald WI, Compston A, Edan G et al (2001) Recommend diagnostic criteria for multiple sclerosis: guidelines from the international panel on the diagnosis of multiple sclerosis. Ann Neurol 50:121–127
Polman CH, Reingold SC, Edan G et al (2005) Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann Neurol 58:840–846
Wattjes MP, Harzheim M, Kuhl CK et al (2006) Does high-field MR imaging have an influence on the classification of patients with clinically isolated syndromes according to current diagnostic mr imaging criteria for multiple sclerosis? AJNR Am J Neuroradiol 27:1794–1798
Acknowledgments
The authors wish to express their gratitude to Dr. Dirk L. Knol for statistical advice, to Dr. John P. Mugler 3rd for his help in developing and programming the sequences and to Dr. Joost P.A. Kuijer for implementing the 3D-DIR sequence. The MS Center Amsterdam and BM were financially supported by the Dutch MS Research Foundation (grant no. 02–358b). This work was also supported in part by funding provided by the US National Multiple Sclerosis Society (RG 3574-A-1), as well as the National Institutes of Health (R01 NS35142).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Moraal, B., Roosendaal, S.D., Pouwels, P.J.W. et al. Multi-contrast, isotropic, single-slab 3D MR imaging in multiple sclerosis. Eur Radiol 18, 2311–2320 (2008). https://doi.org/10.1007/s00330-008-1009-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-008-1009-7