Abstract
The 2016 World Health Organization Classification of Tumors of the Central Nervous System incorporates the use of molecular information into the classification of brain tumors, including grade II and III gliomas, providing new prognostic information that cannot be delineated based on histopathology alone. We hypothesized that these genomic subgroups may also have distinct imaging features. A retrospective single institution study was performed on 40 patients with pathologically proven infiltrating WHO grade II/III gliomas with a pre-treatment MRI and molecular data on IDH, chromosomes 1p/19q and ATRX status. Two blinded Neuroradiologists qualitatively assessed MR features. The relationship between each parameter and molecular subgroup (IDH-wildtype; IDH-mutant-1p/19q codeleted-ATRX intact; IDH-mutant-1p/19q intact-ATRX loss) was evaluated with Fisher’s exact test. Progression free survival (PFS) was also analyzed. A border that could not be defined on FLAIR was most characteristic of IDH-wildtype tumors, whereas IDH-mutant tumors demonstrated either well-defined or slightly ill-defined borders (p = 0.019). Degree of contrast enhancement and presence of restricted diffusion did not distinguish molecular subgroups. Frontal lobe predominance was associated with IDH-mutant tumors (p = 0.006). The IDH-wildtype subgroup had significantly shorter PFS than the IDH-mutant groups (p < 0.001). No differences in PFS were present when separating by tumor grade. FLAIR border patterns and tumor location were associated with distinct molecular subgroups of grade II/III gliomas. These imaging features may provide fundamental prognostic and predictive information at time of initial diagnostic imaging.
Similar content being viewed by others
Introduction
WHO grade II and III cerebral gliomas, also recently termed lower grade gliomas (LGGs), comprise a heterogeneous group of infiltrative neoplasms with astrocytic and oligodendroglial morphology. These tumors have a wide range of both progression-free survival (PFS) and overall survival (OS); some respond to therapy with OS approaching 15 years, while others rapidly progress to glioblastoma [1]. Traditional histopathological classification and grading, the conventional gold standard, is limited by high intraobserver and interobserver variability. Subsequently, this uncertainty in predicting a tumor’s pathogenesis can complicate the selection of optimal treatment and estimation of a patient’s eventual outcome [2, 3].
During the past several decades, tremendous strides have been made to elucidate the underlying molecular alterations present in gliomas [4]. Codeletion of chromosome arms 1p and 19q (henceforth termed 1p/19q codel) was first established as an association with oligodendroglial morphology; subsequently, it was validated as a prognostic marker and shown to be predictive of response to chemoradiation [5,6,7,8]. Following this, isocitrate dehydrogenase gene mutations (referred to as IDH mutations, and reflecting either a mutation in IDH1 or IDH2) were identified; this (collective) mutation is thought to be an early step in gliomagenesis, and estimated to occur in 70–90% of grade II/III gliomas. IDH mutations have also been well-validated as both prognostic and predictive markers of prolonged survival relative to IDH-wildtype tumors [9,10,11,12,13,14]. More recently, explorations into mutations of the alpha-thalassemia/ mental retardation syndrome X-linked gene (termed ATRX), revealed that its loss is more common in astrocytomas, and that it is mutually exclusive with 1p/19q codel [15,16,17]. Furthermore, initial studies suggest it may be associated with a favorable prognosis [17, 18]. Similarly, TP53 mutations are also more frequent in astrocytomas, a marker of clinical outcome and mutually exclusive with 1p/19q codel, thereby commonly co-existing with ATRX mutations [1].
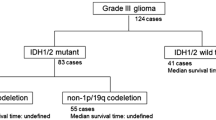
Adding momentum to this field, two recent studies have demonstrated that molecular characterization of glioma including IDH and 1p/19q status is more robust for prediction of clinical outcomes, compared with histological classification. In particular, it was determined that IDH-wildtype gliomas resemble aggressive grade IV gliomas clinically and molecularly, suggesting these should be termed “stealth glioblastomas” [1, 4]. Recently, these efforts were validated in the 2016 World Health Organization Classification of Tumors of the Central Nervous System in which molecular parameters were integrated with histopathology into glioma characterization to create more biologically homogenous groups and improve predictive and prognostic accuracy [19]. As a complement, large-scale sequencing studies have helped define three groups of clinically and biologically distinct gliomas delineated by IDH mutation with 1p/19q codeletion (and TERT promoter mutation), IDH mutation with TP53 mutation and frequent ATRX mutation, and IDH-wildtype status (with TERT promoter mutation and glioblastoma-associated genomic changes) [20]. Subsequently, following from these collective guidelines, we classified grade II/III gliomas into 3 distinct molecular subgroups: IDH-wildtype (IDH-WT), IDH-mutant-1p/19q codeleted-ATRX intact (IDH-MT1p/19qcodel) and IDH-mutant-1p/19q intact-ATRX loss (IDH-MT ATRXloss), with the final group also characteristically having a TP53 mutation.
While molecular analyses can distinguish these subgroups, difficulties in creating uniform genotype testing and reporting can limit its utility, and the technique is also dependent on tissue sampling [21]. MRI is the primary modality for initial diagnostic work-up of gliomas prior to invasive biopsy. As such, distinct imaging features could be defined on MRI reflecting the subgroups underlying molecular differences and yielding essential predictive and prognostic information at the initial diagnostic MRI to help guide clinical decisions. While previous studies have demonstrated that IDH and 1p/19q status have some distinct imaging features in selective subsets of gliomas, there is a dearth of studies evaluating these relationships collectively including both grades. In this study, we hypothesized that these aforementioned grade II/III glioma subgroups, devised from the 2016 WHO classification and large-scale sequencing studies, have unique imaging signatures that may aid in the prediction of patient outcomes.
Methods
Patient cohort
A single institution retrospective study was performed at the Moores Cancer Center at the University of California, San Diego with institutional review board approval. From April 2008 to March 2015, 57 patients were identified who had a pathologically proven grade II or III glioma. Patients were excluded for lack of pre-treatment imaging (defined as prior to surgical resection or initiation of chemoradiotherapy), incomplete imaging (defined as incomplete pre- or post-contrast, FLAIR, ADC and/or DWI sequences), incomplete molecular data (IDH, 1p/19q, and ATRX status) or tumor upstaging to glioblastoma. Upon neuroradiologist review of the cases, one patient was excluded for having a collision lesion: a concomitant arteriovenous malformation and glioma. The final cohort consisted of 40 patients. The specifics are delineated in the STARD chart (Online Resource 1).
Molecular analysis
1p/19q status was determined by either OncoScan microarray analysis (Affymetrix, Santa Clara, CA) or fluorescence in situ hybridization (FISH) performed on formalin fixed paraffin embedded tissue. The majority of IDH and all ATRX status were analyzed by whole exome next-generation sequencing (Illumina, San Diego, CA), as previously described by Dubbink et al. [22]. A minority of IDH status was determined by mutation-specific immunohistochemistry (IHC) for the most common IDH1 mutation (R132H). Following this, the tumors were then classified into three subgroups: Group (1) IDH-WT; Group (2) IDH-MT1p/19qcodel and Group (3) IDH-MT ATRXloss.
Imaging technique and analysis
All of the pre-treatment MR scans were acquired on either a 1.5T or 3T MR unit and included T1 pre- and post-contrast sequences, FLAIR, and DWI/ADC. The exact scanning parameters were variable as some of the scans were performed at outside institutions prior to the patient’s referral to our center. However, all scans were deemed to be of diagnostic quality without significant artifacts and included at least T1W pre- and post-contrast, FLAIR, ADC and DWI sequences. Of the initial cohort, only five scans had perfusion data available.
All MRIs were jointly reviewed by two board-certified neuroradiologists (N.F. with 6 and J.H. with 10 years of post-graduate experience) who were blinded to the clinical history, histopathologic diagnosis, and molecular status. Imaging characteristics were qualitatively evaluated and scored based on previously agreed-upon scales for FLAIR border, contrast enhancement, and diffusion characteristics. A consensus was reached for each category on all 40 patients (Fig. 1). The MR imaging features were based on whole tumor volume and included: (1) tumor location (frontal, parietal, temporal, occipital, multilobar, or cerebellar); (2) frontal only versus non-frontal/involving more than the frontal lobe (based on prior literature demonstrating a frontal lobe predominance of IDH mutant and 1p/19q codeleted grade II/III gliomas); (3) hemisphere (right, left or bilateral); (4) tumor border definition on FLAIR (well-defined: where the border between tumor and normal appearing white matter can be delineated; ill-defined: where the border between tumor and normal appearing white matter is hazy; cannot be defined: where the border between tumor and normal appearing white matter cannot be delineated); (5) volume of contrast enhancement (<25, 25–75 or >75%); and (6) qualitative assessment of diffusion on DWI/ADC (restricted: hyperintense on DWI and hypointense on ADC) [23]. The imaging classification scheme is demonstrated in Fig. 1.
The top panel demonstrates the classification for FLAIR tumor border: the image on the left shows a left frontal tumor with a well-defined border; the middle image shows a left frontal tumor classified as an ill-defined border; the right image shows a left sided multilobar tumor whose border cannot be discerned. The bottom panel demonstrates the classification for degree of tumor contrast enhancement: the image on the left shows a left sided multilobar tumor with no contrast enhancement; the middle image shows a left frontal tumor in which >25% but <50% of the tumor bulk enhances; the right sided image shows a multilobar glioblastoma in which >50% of the tumor bulk enhances (no tumors in this study cohort had this characteristic)
Progression free survival (PFS) analysis
PFS was calculated from the date of tissue diagnosis to the date of documented imaging progression. Tumor progression was defined as unequivocal progression on MRI in conjunction with the assessment of the treating neuro-oncologist. Patients without documented progression were censored at the date of the last stable MRI scan.
Statistical analysis
Analysis of variance (ANOVA) and chi square tests were performed to test for group differences in age and gender distribution. Fisher’s exact test was used to evaluate the relationship between imaging characteristics and molecular status. Analysis was initially performed comparing all three groups. Subsequently, to elucidate potential inter-group differences, analysis was performed to compare the collective IDH-MT groups versus the IDH-WT group and the two IDH-MT groups.
Univariate Cox Proportional Hazard (CPH) models and Kaplan–Meier analyses were conducted to assess the contribution of age, gender, and type of surgery (gross total resection vs. subtotal resection vs. biopsy) to PFS. Significant univariate predictors were included as covariates in the main Kaplan Meier analysis, which was performed to assess for differences in PFS among the molecular subgroups and compared using a log-rank test. PFS analysis was also performed based on grade across all three molecular subtypes. (Statistical Package for the Social Sciences Statistics, Version 21. Armonk, New York).
Results
Cohort characteristics, molecular subgroups, and tumor grade
The clinical characteristics of the final cohort are summarized in Online Resource 2.
A significant difference existed in age among the three subgroups [F (2,39) = 7.16, p = 0.002]. A post-hoc analysis revealed that the IDH-MT ATRXloss patients (mean age = 35.4 years) were significantly younger than both the IDH-MT1p/19qcodel (mean age = 46.6 years; p = 0.046) and the IDH-WT (mean age = 51.6 years; p = 0.002) patients. The overall cohort consisted of 13 (32.5%) females and 27 (67.5%) males. The gender breakdown was also statistically significant (p = 0.035) with more males in the IDH-WT group (92.3%) versus the two MT groups (IDH-MT1p/19qcodel: 66.7%, IDH-MT ATRXloss: 46.7%). CPH analysis demonstrated that age was not associated with PFS (95% CI 0.969–1.039, DF 1, p = 0.866). Additionally, gender was not associated with PFS (Log-Rank: Chi-Square 1.938, DF 1, p = 0.164). However, type of surgery was associated with PFS (Log-Rank:Chi-Square 6.9, DF 2, p < 0.05).
MR imaging correlates of molecular subgroups
The radiologic characteristics of the cohort are summarized in Table 1. Tumor location was associated with the molecular subgroup, in that the IDH-MT tumors were predominantly located in the frontal lobe (p = 0.006). There was no significant difference in location between the IDH-MT groups.
The tumor border on FLAIR was also highly associated with the molecular subgroup. IDH-WT tumors were more likely to have a non-definable border, while the collective IDH-MT tumors had well-defined or slightly ill-defined borders (p = 0.019). Specifically, of the 27 IDH-MT tumors, only one had a border that could not be defined, whereas this was true for six of the 13 IDH-WT tumors. There was no significant difference in border pattern between the two IDH-MT groups (p = 0.684).
There was no significant difference in contrast enhancement among the three groups (p = 0.336). In fact, none of the tumors (all grade II/III) in this cohort demonstrated marked enhancement. Only five (12.5%) of the tumors demonstrated restricted diffusion. This included two in the WT and three in the MT groups. This was not significantly different across the three groups (p = 0.345). Table 1 demonstrates the MRI features of the cohort according to the molecular subgroup. Figures 2 and 3 compare imaging between IDH-MT and IDH-WT grade II and III tumors respectively.
The top panel demonstrates MR images from a patient with a left-sided frontal lobe IDH-MT1p/19qcodel oligodendroglioma (WHO grade II) who did not experience progression during the study. The bottom panel demonstrates MR images from a right-sided multilobar IDH-WT diffuse astrocytoma (also WHO grade II) who progressed within 8 months following tissue sampling. The mutant tumor has a well-defined border on FLAIR whereas the wildtype tumor border cannot be delineated (arrows). Both tumors have minimal (<25%) contrast enhancement. Neither tumor demonstrates restricted diffusion
The top panel demonstrates MR images from a patient with a right-sided frontal lobe IDH-MT ATRXloss anaplastic astrocytoma (WHO grade III) who did not experience progression during the study. The bottom panel demonstrates MR images from a bilateral multilobar IDH-WT anaplastic astrocytoma (also WHO grade III) who progressed less than 14 months following tissue sampling. The mutant tumor has a well-defined border on FLAIR whereas the wildtype tumor border cannot be delineated. Both tumors have minimal (<25%) contrast enhancement. Neither tumor demonstrates restricted diffusion. H&E sections of the two tumors show relatively similar morphology; however, the mutant tumor stains positive for an antibody against IDH confirming the mutation presence, which is negative in the IDH-WT tumor
PFS
The median PFS within the IDH-WT subgroup was 8 months with 77% (n = 10) progressing during the study duration, which was significantly shorter compared to the IDH-MT subgroups (Log-Rank: Chi-Square 40.1, DF 5, p < 0.001, Fig. 4). Among the IDH-MT subgroups, the IDH-MT1p/19qcodel had a median PFS of 51 months with 42% (n = 5) progressing during the study and the IDH-MT ATRXloss had a median PFS of 43 months with 40% (n = 6) of patients progressing (Fig. 4). There was no difference in PFS between the two IDH-MT groups. Further analysis revealed no significant differences in PFS among the whole cohort when based on grade (II or III) (Log-Rank: Chi-Square = 7.3, DF 5, p = 0.202).
Kaplan–Meier curve with log-rank tests were used to examine differences in PFS among the different molecular subgroups with type of surgery as a covariate. The red line represents the IDH-WT tumors (n = 13), the blue line represents the IDH-MT1p/19qcodel tumors (n = 12), and the green line represents the IDH-MT ATRXloss tumors (n = 15). The median PFS in the IDH-WT subgroup was significantly shorter (8 months) than PFS in the IDH-MT1p/19qcodel (51 months) or the IDH-MT ATRXloss (43 months: p < 0.001) subgroup. However there was no difference in PFS between the two IDH MT subgroups
During the duration of follow up, six of the patients in the cohort died and one went to hospice. Of these seven patients, six were in the IDH-WT group.
Discussion
The delineation of three distinct molecular subgroups of grade II/III gliomas (IDH-WT, IDH-MT1p/19qcodel and IDH-MT ATRXloss) validates years of research linking 1p/19q and IDH status with clinical outcome [6, 10]. Indeed, molecular status can yield more prognostic information than traditional histopathology alone. The novel integration of genotypic information with tumor histology in the 2016 WHO Classification of Tumors of the CNS attests to the significant role radiogenomics can play within the field of neuro-oncology.
Using primarily next-generation sequencing techniques and OncoScan microarray analysis to determine three distinct molecular subgroups of grade II/III gliomas, we demonstrate that on FLAIR imaging, the IDH-MT tumors had more well-defined borders in contrast to the IDH-WT tumors whose borders could not be defined. Given that IDH-WT tumors are known to be clinically and molecularly similar to glioblastomas, it follows that these tumors show a more infiltrative pattern on MRI. This characteristic FLAIR pattern could help distinguish the sentinel molecular classification of a tumor at initial imaging diagnosis.
This differentiating FLAIR border adds to prior work of other investigators including Metellus et al. who found that 100% of IDH-WT tumors in their cohort had indistinct borders in contrast to only 45% of IDH-MT tumors. However, their study was limited to only grade II gliomas and used PCR for assessment of IDH mutations [23]. Qi et al. found similar results, but only evaluated astrocytomas [24]. Our study expanded on these by including grade II and III astrocytomas and oligodendrogliomas; furthermore, employing predominantly a next-generation sequencing technique allowed us to capture rare IDH mutations (including non-R132H IDH1 and all IDH2 mutations) enabling us to more accurately characterize our sample [25]. In fact, less sensitive methods may have hampered classification in previous studies. We did not find a difference in the FLAIR border characteristics between the two IDH mutant groups. This is in contrast to Jenkinson et al., who in 2006 prior to the 2016 WHO guidelines, investigated the differences in tumor borders of histologically classified oligodendroglial tumors and found that 1p/19q intact tumors had sharp/smooth borders while the 1p/19q codel tumors rarely had sharp borders [26].
Our findings that the IDH-MTs collectively had a frontal lobe predilection, echoes previous studies [24, 27]. The current hypothesis is that the IDH-MT tumors arise from a neural precursor population with a defined spatial and temporal location, and since the IDH mutation precedes 1p/19q codeletion in tumorgenesis, it follows that the 1p/19q codeleted tumors are also frontal lobe predominant [11, 28, 29]. Similarly, as we did not find any substantive differences between the IDH-MT1p/19qcodel and the IDH-MT ATRXloss groups, this may be secondary to their common origin path in gliomagenesis (IDH mutation), and more advanced imaging techniques may be needed to elucidate their differences.
Our study is in accordance with prior publications by demonstrating the prognostic significance of these novel molecular subgroups [30]. We found that median PFS was significantly shorter in the IDH-WT group compared to both IDH-MT groups (p < 0.001); whereas, there was no difference in PFS between the two IDH-MT groups. While OS was not a primary endpoint evaluated in this study, it should be noted that of the seven patients who died or went to hospice, six were in the IDH-WT group. Importantly, there were no significant differences in PFS among the three groups when based on grade (II or III). This reinforces prior studies that found no significant prognostic differences between IDH-MT grade II and III tumors [31, 32]. In fact, Suzuki et al. stated that grade III IDH-MT tumors showed OS comparable to corresponding grade II tumors and recommended managing them as low-grade tumors [33]. Additionally, the significant difference in age among the three subgroups (with the IDH-WT being older than the collective IDH-MT groups) is in accordance with Eckel-Pastow et al. and should be expected since IDH-WT tumors are on the spectrum of glioblastomas, which tend to present later in life than less aggressive gliomas [4].
While genotyping is presently the gold standard for classifying patients into different molecular subgroups with established predictive and prognostic value, radiogenomic delineation can add considerable value. First, these genotyping techniques as well as surrogate immunohistochemistry may not be readily available at all institutions and the variability among genotyping assays may lead to imperfect groups when less sensitive modalities are used. Furthermore, certain tumors such as brainstem gliomas may not be amenable to biopsy, and if sampling is attempted, intertumoral regional heterogeneity is a potential problem with possible exclusion of tumor cell subpopulations [33]. In contrast to these shortcomings, imaging features, specifically FLAIR border pattern, can be readily evaluated on the initial diagnostic MRI, potentially providing predictive and prognostic information. The ability to differentiate IDH-mutant and IDH-wildtype tumors at the time of initial imaging may influence decisions regarding the extent and strategy of surgical resection. Additionally, FLAIR border analysis may play a pertinent role in monitoring patients, specifically in assessing for a change in molecular phenotype with progression or recurrence.
While other investigators have found similar associations between IDH-status and FLAIR border pattern, our cohort is unique in that we examined both grade II and III gliomas and we devised groups prescribed by the recently published 2016 WHO Consensus and complementary large-scale sequencing studies, particularly incorporating IDH, 1p/19q and ATRX status. Additionally, we expanded the present knowledge base by assessing for differences among these groups utilizing several common imaging techniques (FLAIR, contrast-enhancement and diffusion) thereby emulating tumor evaluation in daily clinical neuroradiology practice. Although the current clinical utility of these imaging-genomic associations may be primarily reserved for cases where biopsy cannot be performed, given the rapidly evolving field of radiogenomics, these associations may have much more significant implications in the near future such as in vivo assessment of treatment response versus tumor progression.
There are limitations of this study based on its retrospective nature. Additionally, there was heterogeneity in the MRI acquisition parameters, since several of the scans were from outside institutions due to the need to assess pre-treatment MRIs. This precluded performing a formal quantitative assessment of imaging characteristics. However, this is in line with routine clinical practice, which utilizes qualitative analysis of lesions for initial assessment as well as follow-up examinations. It is possible that future studies could incorporate quantitative analysis to further distinguish molecular subgroups. Furthermore, while we accounted for type of surgery in our Kaplan Meier curves, chemotherapy regimens were varied among the groups, which could have impacted PFS (perhaps limiting our ability to find groups differences between the IDH-MT subgroups). However, all of the evaluated MRIs were pre-treatment and therefore the imaging analysis was not affected by treatment heterogeneity.
Conclusion
In this study we found that imaging features, particularly FLAIR border pattern and tumor location, could distinguish molecular subgroups of grade II and III gliomas based on the Updated WHO Consensus Classification, and the ability to distinguish these subgroups on initial diagnostic imaging may impact clinical decision making. Furthermore, these molecular subgroups provide important prognostic information that is not captured by histological classification and grading alone. Future studies should evaluate whether these imaging features can characterize progressive or recurrent disease, and guide therapeutic decisions including surgical and radiotherapy strategies.
Change history
13 September 2017
In the initial online publication, the values in the last two rows in Table 1 were in the wrong rows. The original article has been corrected.
References
The Cancer Genome Atlas Research Network (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372:2481–2498
Coons SW, Johnson PC, Scheithauer BW et al (1997) Improving diagnostic accuracy and interobserver concordance in the classification and grading of primary gliomas. Cancer 79:1381–1393
Van den Bent MJ (2010) Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician’s perspective. Acta Neuropathol 120:297–304
Eckel-Passow JE, Lachance DH, Molinaro AM et al (2015) Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372:2499–2508
Reifenberger J, Reifenberger G, Liu L et al (1994) Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol 145:1175–1190
Jenkins RB, Blair H, Ballman KV et al (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res 66:9852–9861
Cairncross G, Wang M, Shaw E et al (2013) Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol 31:337–343
Van den Bent MJ, Brandes AA, Taphoorn MJ et al (2013) Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol 31:344–350
Sanson M, Marie Y, Paris S et al (2009) Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 27:4150–4154
Yan H, Parsons DW, Jin G et al (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773
Watanabe T, Nobusawa S, Kleihues P et al (2009) IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 174:1149–1153
Hartmann C, Meyer J, Balss J et al (2009) Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol 118:469–474
Duncan CG, Barwick BG, Jin G et al (2012) A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res 22:2339–2355
Noushmehr H, Weisenberger DJ, Diefes K et al (2010) Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17:510–522
Liu XY, Gerges N, Korshunov A et al (2012) Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol 124:615–625
Cairncross JG, Wang M, Jenkins RB et al (2014) Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. J Clin Oncol 32:783–790
Kannan K, Inagaki A, Silber J et al (2012) Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget 3:1194–1203
Wiestler B, Capper D, Holland-Letz T et al (2013) ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol 126:443–451
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (2016) WHO classification of tumors of the central nervous system. IARC, Lyon
Louis DN, Perry A, Burger P et al (2014) International society of Neuropathology-haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol 24:429–435
Dubbink HJ, Atmodimedjo PN, Kros JM et al (2016) Molecular classification of anaplastic oligodendroglioma using next-generation sequencing: a report of the prospective randomized EORTC Brain Tumor Group 26951 phase III trial. Neuro Oncol 18:388–400
Metellus P, Coulibaly B, Colin C et al (2010) Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol 120:719–729
Qi S, Yu L, Li H et al (2014) Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol Lett 7:1895–1902
da Fonseca AJ, Galvão RS, Miranda AE et al (2016) Comparison of three human papillomavirus DNA detection methods: next generation sequencing, multiplex-PCR and nested-PCR followed by Sanger based sequencing. J Med Virol 5:888–894
Jenkinson MD, du Plessis DG, Smith TS et al (2006) Histological growth patterns and genotype in oligodendroglial tumors: correlation with MRI features. Brain 129:1884–1891
Reyes-Botero G, Caroline D, Idbaih A et al (2014) Contrast enhancement in 1p/19q-codeleted anaplastic oligodendrogliomas is associated with 9p loss, genomic instability and angiogenic gene expression. Neuro Oncol. 16:662–670
Lai A, Kharbanda S, Pope WB et al (2011) Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol 29:4482–4490
Persson AI, Petritsch C, Swartling FJ et al (2010) Non-stem cell origin for oligodendroglioma. Cancer Cell 18:669–682
Leeper HE, Caron AA, Decker PA et al (2015) IDH mutation, 1p19q codeletion and ATRX loss in WHO grade II gliomas. Oncotarget 6:30295–30305
Reuss DE, Mamatjan Y, Schrimpf D et al (2015) IDH mutant diffuse and anaplastic astrocytomas have similar age at presentation and little difference in survival: a grading problem for WHO. Acta Neuropathol 129:867–873
Olar A, Wani KM, Alfaro-Munoz KD et al (2015) IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II–III diffuse gliomas. Acta Neuropathol 129:585–596
Suzuki H, Aoki K, Chiba K et al (2015) Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet 47:458–466
Acknowledgements
We would like to thank patients at the UCSD Moores Cancer Center Neuro-Oncology Program for their generous participation.
Funding
National Institutes of Health Grants R01NS065838 (C.R.M.), RC2 DA29475 and EB00790-06 (A.M.D.), UL1TR000100 and KL2TR00099 (J.A.H.); National Cancer Institute Cancer Center Specialized Grant P30CA023100 (C.R.M.); National Science Foundation Grant 1430082 (N.S.W.); American Cancer Society Award ACS-IRG 70-002 (J.A.H.); and American Cancer Society Research Scholar Grant RSG-15-229-01-CCE (C.R.M.).
Author information
Authors and Affiliations
Corresponding author
Additional information
The original version of this article has been revised: The values in the last two rows in Table 1 have been switched to the correct rows.
An erratum to this article is available at https://doi.org/10.1007/s11060-017-2620-8.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Delfanti, R.L., Piccioni, D.E., Handwerker, J. et al. Imaging correlates for the 2016 update on WHO classification of grade II/III gliomas: implications for IDH, 1p/19q and ATRX status. J Neurooncol 135, 601–609 (2017). https://doi.org/10.1007/s11060-017-2613-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-017-2613-7