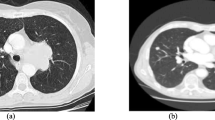
Abstract
Purpose
This study aims to determine the three-dimensional (3D) texture features extracted from intensity and high-order derivative maps that could reflect textural differences between bladder tumors and wall tissues, and propose a noninvasive, image-based strategy for bladder tumor differentiation preoperatively.
Methods
A total of 62 cancerous and 62 wall volumes of interest (VOI) were extracted from T2-weighted MRI datasets of 62 patients with pathologically confirmed bladder cancer. To better reflect heterogeneous distribution of tumor tissues, 3D high-order derivative maps (the gradient and curvature maps) were calculated from each VOI. Then 3D Haralick features based on intensity and high-order derivative maps and Tamura features based on intensity maps were extracted from each VOI. Statistical analysis and recursive feature elimination-based support vector machine classifier (RFE-SVM) was proposed to first select the features with significant differences and then obtain a more predictive and compact feature subset to verify its differentiation performance.
Results
From each VOI, a total of 58 texture features were derived. Among them, 37 features showed significant inter-class differences (\(P\le 0.01\)). With 29 optimal features selected by RFE-SVM, the classification results namely the sensitivity, specificity, accuracy and area under the curve (AUC) of the receiver operating characteristics were 0.9032, 0.8548, 0.8790 and 0.9045, respectively. By using synthetic minority oversampling technique to augment the sample number of each group to 200, the sensitivity, specificity, accuracy an AUC value of the feature selection-based classification were improved to 0.8967, 0.8780, 0.8874 and 0.9416, respectively.
Conclusions
Our results suggest that 3D texture features derived from intensity and high-order derivative maps can better reflect heterogeneous distribution of cancerous tissues. Texture features optimally selected together with sample augmentation could improve the performance on differentiating bladder carcinomas from wall tissues, suggesting a potential way for tumor noninvasive staging of bladder cancer preoperatively.









Similar content being viewed by others
References
American Cancer Society (2015) Cancer facts and figures 2015. American Cancer Society, Atlanta, pp 8–16
Torre L, Bray F, Siegel R, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics 2012. CA Cancer J Clin 65(2):87–108
National Comprehensive Cancer Network (2015) NCCN clinical practice guidelines in oncology, pp 30–33
Stein J, Lieskovsky G, Cote R, Groshen S, Feng A, Boyd S, Skinner E, Bochner B, Thangathurai D, Mikhail M, Raghavan D, Skinner D (2001) Radical cystectomy in the treatment of invasive bladder cancer: long-term results in 1,054 patients. J Clin Oncol 19(3):666–675
Makram M, Michaël P, Marc Z, Djillali S, Bernard D (2003) The value of a second transurethral resection in evaluating patients with bladder tumours. Eur Urol 43(3):241–245
Jakse G, Algaba F, Malmstrom P, Oosterlinck W (2004) A second-look TUR in T1 transitional cell carcinoma: why? Eur Urol 45(5):539–546
Kim B, Semelka R, Ascher S, Chalpin D, Carroll P, Hricak H (1994) Bladder tumor staging: comparison of contrast-enhanced CT, Ti- and T2-weighted MR Imaging, dynamic gadolinium-enhanced imaging, and late gadolinium-enhanced imaging. Radiology 193:239–245
Xiao D, Zhang G, Liu Y, Yang Z, Zhang X, Li L, Jiao C, Lu H (2016) 3D detection and extraction of bladder tumors via MR virtual cystoscopy. Int J Comput Assist Radiol Surg 11(1):89–97
Rais-Bahrami S, Pietryga J, Nix J (2015) Contemporary role of advanced imaging for bladder cancer staging. Urol Oncol 18(2):168–177
Shi Z, Yang Z, Zhang G, Cui G, Xiong X, Liang Z, Lu H (2013) Characterization of texture features of bladder carcinoma and the bladder wall on MRI: initial experience. Acad Radiol 20(8):930–938
Zhang X, Liu Y, Yang Z, Tian Q, Zhang G, Xiao D, Cui G, Lu H (2015) Quantitative analysis of bladder wall thickness for magnetic resonance cystoscopy. IEEE Trans Biomed Eng 62(10):2402–2409
Zhao Y, Liang Z, Zhu H, Han H, Duan C, Yan Z, Lu H, Gu X (2013) Bladder wall thickness mapping for magnetic resonance cystography. Phys Med Biol 58(15):5173–5192
Ganeshan B, Miles K, Young R, Chatwin C (2009) Texture analysis in non-contrast enhanced CT: impact of malignancy on texture in apparently disease-free areas of the liver. Eur J Radiol 70:101–110
Ng F, Ganeshan B, Kozarski R, Miles K, Goh V (2013) Assessment of primary colorectal cancer heterogeneity by using whole-tumor texture analysis: contrast-enhanced CT texture as a biomarker of 5-year survival. Radiology 266(1):177–184
Sheshadri H, Kandaswamy A (2007) Experimental investigation on breast tissue classification based on statistical feature extraction of mammograms. Comput Med Imag Graph 31:46–58
Fu J, Yu Y, Lin H, Chai J, Chen CC (2014) Feature extraction and pattern classification of colorectal polyps in colonoscopic imaging. Comput Med Imag Graph 38(4):267–275
Bayanati H, Thornhill E, Souza C, Virmani V, Gupta A, Maziak D, Amjadi K, Dennie C (2015) Quantitative CT texture and shape analysis: can it differentiate benign and malignant mediastinal lymph nodes in patients with primary lung cancer? Eur Radiol 25(2):480–487
Song B, Zhang G, Lu H, Wang H, Zhu W, Pickhardt P, Liang Z (2014) Volumetric texture features from higher-order images for diagnosis of colon lesions via CT colonography. Int J Comput Assist Radiol Surg 9(6):1021–1031
Hu Y, Liang Z, Song B, Han H, Pickhardt P, Zhu W, Duan C, Zhang H, Barish M, Lascarides C (2016) Texture feature extraction and analysis for polyp differentiation via computed tomography colonography. IEEE Trans Med Imag 35(6):1522–1531
Fetit A, Novak J, Peet A, Arvanitits T (2015) Three-dimensional textural features of conventional MRI improve diagnostic classification of childhood brain tumours. NMR Biomed 28(9):1174–1184
Nailon W, Redpath A, McLaren D (2008) Characterisation of radiotherapy planning volumes using textural analysis. Acta Oncol 47(7):1303–1308
Xu X, Zhang X, Tian Q, Q Tian Q, Zhang G, Lu H (2016) Differentiating bladder carcinoma from bladder wall using 3D textural features: an initial study. SPIE Med Image Process 2016:1–11
Simoes R, Walsum A, Slump C (2014) Classification and localization of early-stage Alzheimer’s disease in magnetic resonance images using a patch-based classifier ensemble. Neuroradiology 56(9):1–12
Zhang G, Song B, Zhu H, Liang Z (2012) Computer-aided diagnosis in CT colonography based on bi-labeled classifier. Int J CARS 7(Suppl):S274
Han F, Wang H, Zhang G, Han H, Song B, Li L, Moore W, Lu H, Zhao H, Liang Z (2015) Texture feature analysis for computer-aided diagnosis on pulmonary nodules. J Digit Imag 28(1):99–115
Haralick R, Shanmugan K, Dinstein I (1973) Texture features for image classification. Trans Syst Man Cybern SMC–3(6):610–621
Majtner T, Svoboda D (2012) Extension of tamura texture features for 3D fluorescence microscopy. Second international conference on 3D Imaging, pp 301–307
Tamura H, Mori S, Yamawaki T (1978) Texture features corresponding to visual perception. IEEE Trans Syst Man Cybern SMC–8(6):460–473
Zou J, Ji Q, Nagy G (2007) A comparative study of local matching approach for face recognition. IEEE Trans Image Process 16(10):2617–2628
Zacharaki E, Wang S, Chawla S, Yoo D, Wolf R, Melhem E, Davatzikos C (2009) Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 62(6):1609–1618
Zyout I, Czajkowska J, Grzegorzek M (2015) Multi-scale textural feature extraction and particle swarm optimization based model selection for false positive reduction in mammography. Comput Med Imag Graph 46:95–107
Laimighofer M, Krumsiek J, Buettner F, Theis F (2016) Unbiased prediction and feature selection in high-dimensional survival regression. J Comput Biol 23(4):279–290
Guyon I, Weston J, Barnhill S, Vapnik V (2002) Gene selection for cancer classification using support vector machines. Mach Learn 46:389–422
Rakotomamonjy A (2003) Variable selection using SVM-based criteria. J Mach Learn Res 3:1357–1370
Fehr D, Veeraraghavan H, Wibmer A, Gondo T, Matsumoto K, Vargas H, Sala E, Hricak H, Deasy J (2015) Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. PNAS 112(46):E6265–E6273
Chang C, Lin C (2001) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2(27):1–27
Chawla N, Bowyer K, Hall L, Kegelmeyer W (2002) SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 16(2002):321–357
Lerski R, Straughan K, Schad L, Boyce D, Blüml S, Zuna I (1993) MR image texture analysis—an approach to tissue characterization. Magn Reson Imag 11(6):873–887
Isabelle G, Elisseeff A (2003) An introduction to variable and feature selection. J Mach Learn Res 3:1157–1182
Anaissi A, Goyal M, Catchpoole D, Braytee A, Kennedy P (2016) Ensemble feature learning of genomic data using support vector machine. PLoS ONE 11(6):e0157330
Fang Y, Wang Y, Zhu Q, Wang J, Li G (2016) In silico identification of enhancers on the basis of a combination of transcription factor binding motif occurrences. Sci Rep 6:32476
Zarogianni E, Storkey A, Johnstone E, Owen D, Lawrie S (2016) Improved individualized prediction of schizophrenia in subjects at familial high risk, based on neuroanatomical data, schizotypal and neurocognitive features. Schizophr Res S0920–9964(16):30377–303772. doi:10.1016/j.schres.2016.08.027
Acknowledgements
This work was partially supported by the National Nature Science Foundation of China under Grant No. 81230035, and the Shaanxi Provincial Foundation for Social Development and Key Technology under Grant No. 2015SF177, No. 2016SF302. We would like to thank Mr. Long-Biao Cui from Department of Radiology, Xijing Hospital, the Fourth Military Medical University for the inspiration and discussion of the research idea in this study, and Mr. Dan Xiao for his technical support on medical image processing. More importantly, we would like to thank all editors and the anonymous reviewers for their insightful, helpful and thought-provoking suggestions on the quality improvement of this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Xiaopan Xu and Xi Zhang: co-first authors.
Rights and permissions
About this article
Cite this article
Xu, X., Zhang, X., Tian, Q. et al. Three-dimensional texture features from intensity and high-order derivative maps for the discrimination between bladder tumors and wall tissues via MRI. Int J CARS 12, 645–656 (2017). https://doi.org/10.1007/s11548-017-1522-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-017-1522-8