
Machine learning for semiautomated classification of glioblastoma, brain metastasis and central nervous system lymphoma using magnetic resonance advanced imaging
Introduction
Glioblastoma (GB), central nervous system lymphoma (CNSL), and brain metastasis together represent a large proportion of brain tumors encountered in clinical neuro-oncology. GBs comprise 40% to 50% of primary brain tumors in adults, while brain metastases are found in 10% to 30% of adults with a systemic malignancy, of which nearly half of cases appear on imaging to be solitary metastases (1,2). Primary CNSL comprise up to 4% of primary CNS tumors, with an additional small contribution of secondary CNSL (3,4).
Differentiating these entities may be difficult using conventional magnetic resonance imaging (MRI), as significant potential overlap exists in the degree of post-contrast enhancement and peritumoral FLAIR signal hyperintensity across the 3 tumor classes (5,6). However, establishing the correct diagnosis is important for guiding therapy, as each of these tumor classes carries a different prognosis and requires unique management. GB is an aggressive malignancy generally requiring surgical management and possible adjuvant therapy (7). In the absence of a known malignancy, the diagnosis of a brain metastasis necessitates a metastatic workup to identify the primary disease, which will then guide therapy. Primary CNS lymphoma is generally managed with chemoradiation therapy (8).
The use of advanced MRI including perfusion and diffusion has been investigated for improving the ability to distinguish GB, CNSL, and brain metastasis. For example, CNSL tends to have lower ADC values in comparison to GB and metastasis (9,10), although overlap has been shown (11,12). Perfusion parameters including CBV and permeability measures have also shown promise in differentiating GB, PCL, and metastasis (13-16).
In recent years, machine learning models, including support vector machines (SVMs) and multilayer perceptrons (MLPs), a type of simple neural network, have successfully been used for semi-automated brain tumor classification (17-25). Several of these studies have relied on texture analysis of conventional MR sequences. While other studies have included analysis of perfusion data (17-19,21), the inclusion of permeability parameters has not been commonly reported. The purpose of this study was to investigate whether supervised training of a multiclass SVM or MLP applied to MR perfusion and permeability datasets could reliably differentiate GB, CNSL and brain metastasis using automated feature selection.
Methods
Patients
This retrospective study was conducted between July 2014 and August 2016 according to an approved institutional review board. Inclusion criteria were as follows: (I) histopathologically-proven intracranial GB, CNSL or metastasis and (II) preoperative brain MRI including DSC, dynamic contrast enhanced (DCE) and diffusion weighted imaging (DWI).
Image acquisition
Image acquisition was performed on a 3.0T scanner. DWI was acquired using single-shot spin-echo EPI (TR/TE 4,100/95 ms; FOV: 220 mm × 220 mm; matrix: 128 mm × 128 mm; slices: 30 mm × 5 mm). Diffusion gradients were applied along three orthogonal directions with b=0 and 1,000 s/mm2. DCE perfusion was accomplished using a 3D radial volumetric interpolated examination sequence with the following parameters: TR/TE 4.75/2.2 ms; FA 10°; FOV 220 mm × 220 mm; matrix 256 mm × 192 mm; 30 mm × 5 mm slices with temporal resolution of 6 seconds over a 4-minute acquisition time. Varying flip angle (3°, 5°, and 12°) methodology was implemented for the generation of T1 maps (26). Dynamic susceptibility contrast (DSC) perfusion was performed with a single-shot gradient-echo EPI sequence with the following parameters: TR/TE 1650/30 ms, FA = 90°, FOV 220 mm × 220 mm, matrix 128 mm × 128 mm, 25 mm × 5 mm slices, and 60 dynamic frames.
Image preprocessing
MR perfusion studies were processed using commercially available FDA-approved software (Olea Sphere, Olea Medical SAS, La Ciotat, France). The arterial input function was selected automatically and multiparametric perfusion maps were calculated using an extended Toft model (27) for DCE and block-circulant singular value decomposition technique (28) for DSC. The conventional images (FLAIR and post-contrast T1WI); ADC; CBV and CBF normalized to contralateral white matter (relative CBV and CBF; rCBV and rCBF) from DSC perfusion; and volume transfer constant from extravascular extracellular space (EES) to plasma (K-trans), rate constant from EES to plasma (Kep), EES volume per unit tissue volume (Ve), and blood plasma volume per unit tissue volume (Vp) from DCE perfusion datasets were then exported from the software for subsequent analysis.
The exported images were coregistered to standard Montreal Neurological Institute coordinates by the Functional MRI Software Library (FSL; FMRIB Analysis Group, Oxford, UK, version 5.0) using a 12-degree of freedom transformation and a mutual information cost function (29). This was followed by visual inspection to ensure adequate alignment. Additional preprocessing steps performed in FSL included brain extraction and histogram normalization.
Using a consensus approach, 3-dimensional VOIs were drawn manually on enhancing tumor and peritumoral non-enhancing T2 hyperintensity (NET2) on coregistered T1C+ and FLAIR images, respectively. For patients with more than one tumor, the VOI was confined to the largest lesion. NET2 was defined as the T2 hyperintense region on FLAIR images within 2 cm around the enhancing tumor, excluding necrotic tissue and the enhancing component itself.
For each patient, the T1C+ and NET2 VOIs were applied as inclusion masks to the rCBV, rCBF, K-trans, Kep, Vp, Ve, and ADC maps using FSL to remove all image data outside of the respective VOIs (Figure 1). This generated a total of 14 tumor volumes (7 parameters for each of the 2 VOIs) for each patient.
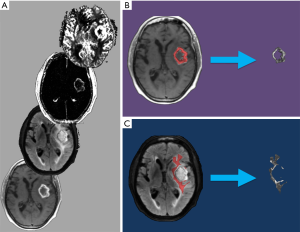
Machine learning
The 14 total T1C+ and NET2 extracted volumes for each patient were then further processed by custom code built by one of the authors (NCS) using the Scikit-learn library v0.18.1 for Python (30). Supervised training of each ML model was accomplished using tumor volume imaging data labeled with the tumor diagnosis. The MLP design utilized a single hidden layer, rectified linear unit activation and an alpha of 0.0001.
Validation of the trained model was performed using a leave-one-subject-out cross-validation structure, described with the following equation:
where K equals the total number of subjects and E equals the error. Leave-one-subject-out entails running K folds, each fold including K-1 subjects for training and the remaining subject held out for validation. True error is then calculated as the average error rate from all K folds.
Separate SVM and MLP models were trained for each head-to-head and three-class tumor volume comparison. For both the SVM and MLP approaches, 56 separate models were trained and validated (14 tumor volumes for each of the 3 head-to-head comparisons and 14 tumor volumes for the three-class comparison), yielding a total of 112 individual trained ML models. For each of the 112 individual tests, a new, “naïve” model was trained. Feature selection was performed de novo automatically by the Scikit-learn module within the nested cross-validation structure to prevent biasing of the trained model that would result from performing feature selection upon the entire subject set. One-versus-rest validation was employed for the three-class comparison.
Trained model total accuracy and receiver operating characteristic data were collected for each of the 112 training and validation cycles.
Subjective interpretation
All imaging studies were reviewed by two board certified neuroradiologists blinded to histopathological diagnosis in independent sessions. Readers were instructed to review all available MR images in each patient and use their best clinical judgment to assign each case with a diagnosis of lymphoma, GB or metastasis.
Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics 24 for Windows (Released 2016; IBM Corp., Armonk, NY, USA). Cohen kappa scores were calculated to quantify the strength of agreement between machine and human interpretation with respect to the ground truth (histopathological diagnosis). For all statistical analysis a P value <0.05 was considered significant.
Results
Twenty-six patients (16 male, 10 female; age 61.8±9.3 years) with glioblastoma (n=9), CNSL (n=8), and metastasis (n=9; lung carcinoma =4, esophageal carcinoma =1, melanoma =1, neuroendocrine carcinoma =1, rectal carcinoma =1, thyroid carcinoma =1) meeting the inclusion criteria were identified.
Classification accuracy
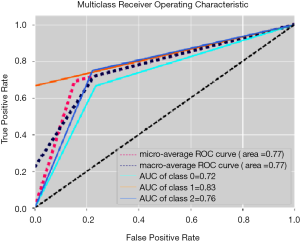
The best performing ML training-validation cycles are included in Table 1. The trained multiclass ML models were able to differentiate the 3 diagnostic classes with a maximum of 69.2% accuracy (kappa 0.540, P=0.01), which was obtained by training an MLP utilizing the Vp values from the perilesional NET2 VOIs (MLP VpNET2). Receiver operating characteristic for this trained MLP VpNET2 model is presented in Figure 2.
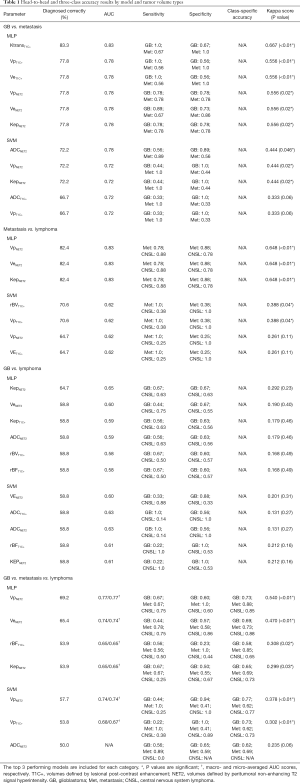
Full table
Head-to-head comparisons for each pair of diagnostic groups demonstrated higher maximum accuracies than the three-class comparisons: 83.3% for GB and metastasis (MLP KtransT1C), 82.4% for metastasis and lymphoma (MLP VpNET2, MLP VeNET2, and MLP KepNET2), and 64.7% for GB and lymphoma (MLP KepNET2).
Human interpretation
Observers A and B identified 17 and 21 out of 26 cases correctly, respectively. The interobserver agreement was k =0.434 (95% CI, 0.167–0.701).
Cohen kappa scores demonstrated the following degrees of inter-rater agreement with respect to histopathological diagnosis: trained multiclass MLP VpNET2 k =0.540 (95% CI, 0.275–0.805), observer A k =0.479 (95% CI, 0.201–0.757), and observer B k =0.712 (95% CI, 0.489–0.934).
Conclusions
The growing interest in machine learning techniques for automated image classification has generated anticipation that this technology may very soon aid radiological diagnosis in clinical practice (31). While many research efforts towards this end have focused on interpretation of conventional CT and MR imaging, the multimodal nature of advanced MRI presents an intriguing target for machine learning experimentation. This study demonstrates that an MLP trained using quantitative perfusion, permeability and diffusion MR imaging can independently differentiate 3 brain tumor classes with a diagnostic accuracy comparable to that of trained neuroradiologists. Additionally, when used in conjunction with human evaluation as a computer-aided diagnosis (CADx) tool the diagnostic yield is increased by approximately 19% over unaided human interpretation.
In this series, the greatest diagnostic accuracy obtained by the ML models in the three-class experiments was achieved by the MLP model trained using VpNET2, which yielded a kappa value of 0.540 (P=0.001) indicating a moderate correlation with the correct histopathological diagnosis. Interestingly, the best accuracy for the multiclass SVC models was also achieved by utilizing the VpNET2 tumor volumes. The Vp parameter (fractional plasma volume) reflects blood plasma volume per unit tissue volume (32) and has shown utility in previous studies for characterizing tumor grade (33) and enabling differentiation of GB and metastasis (34).
The results of this study suggest that differences among the diagnostic classes in the extent of vascularity within the non-enhancing T2 signal hyperintense region surrounding the enhancing tumor component could be used to differentiate the tumor classes (35,36). This is logical, since it is well-known that NET2 surrounding enhancing glioblastoma is likely to represent infiltrative (microscopic) tumor, which may feature neovascularity reflected in the Vp values (37). In contrast, it has been shown that neovascularization is not a prominent histologic feature in CNS lymphoma, which has lower microvascular density as compared with GB (38,39). Therefore, NET2 in lymphoma more likely correlates with densely packed cells with less vascularity as compared with GB and hence lower expected Vp values. NET2 associated with metastatic tumors, which are typically non-infiltrative, is more likely to represent vasogenic edema, which would not be expected to demonstrate elevated vascularity (40,41).
Accuracy results were greater in the head-to-head comparisons than the three-class comparison. This is expected, since narrowing the diagnostic possibilities from 3 to 2 potential diagnoses improves the odds of making a correct classification. However, in the glioblastoma versus lymphoma tests there was a maximum diagnostic accuracy of just 64.7%. The relatively poor ability of the trained models to differentiate glioblastoma and lymphoma within these patients likely also decreased the overall diagnostic accuracies obtained in the three-class tests.
Another possible factor limiting the accuracy obtained in the three-class tests is the use of isolated tumor volumes. This design decision acted as a feature reduction step, “focusing” the model’s attention on the enhancing or NET2 tumor components. It was also an attempt to control for patients with multiple lesions, an imaging feature that if included would have introduced an undesired bias since the purpose of this study was to generate trained models able to differentiate the tumor classes using perfusion and permeability features. However, removing contextual imaging data potentially correlating with a correct diagnosis, such as lesion location, multiplicity, or degree of mass effect on surrounding structures, potentially lowered the diagnostic accuracy of the trained model, disadvantaging it as compared with the human reviewers.
The best accuracy obtained by the trained multiclass ML models was comparable to that of the human reviewers using a simulated real-world clinical workflow that utilized conventional and diffusion-weighted MRI. Further study is required to investigate whether the addition of image texture parameters from conventional MRI in concert with perfusion and permeability parameters may yield ML accuracy superior to that obtained in the current study.
The utility of the trained model may be greater when used as a CADx clinical support tool than as an independent diagnostic tool. Although the diagnostic accuracies of our neuroradiologists were 65% and 81%, respectively, there was a relatively low interobserver agreement between the two readers (16 of 26 cases; k =0.434). Used as a tie-breaker, our best-performing multiclass model resulted in the correct identification of 5 additional cases (19%).
A strength of this investigation is that all patients underwent histopathologic sampling to confirm the diagnosis prior to inclusion. This is crucial since the very purpose of the study is to investigate techniques for differentiating tumors with potentially overlapping imaging features. An additional strength is that feature selection utilized for model training was performed de novo within each cross-validation fold to minimize the risk of biasing and better approximate the performance of the trained model in clinical practice. Some previously described techniques for training ML models to perform multiclass tumor discrimination have pooled best-performing features from head-to-head classifications for subsequent use in multiclass models (17,42). This approach was avoided in the present study because of the risk that features previously extracted from the test subject at hand may be utilized by the trained model, introducing bias and subverting attempts at blinded validation.
Some earlier studies investigating ML for multiclass tumor diagnosis reported high accuracies utilizing developer-specified features such as tumor location, ring enhancement, or hemorrhage (23). This approach was avoided in the current study in favor of using automated feature extraction for several reasons. As opposed to conventional images, perfusion and permeability imaging data are less easily definable in terms of qualitative features. Additionally, the reliance on hard-coded rules may yield a trained model that excels in diagnosing “classic” representations of a given tumor class but struggles with outlier cases in which it is most likely to be of clinical value. Furthermore, automated feature extraction has the significant advantage of scalability when new data are subsequently added to the training set for model refinement.
A potential limitation of this study is the use of manual as opposed to fully automated tumor segmentation, which despite efforts to standardize an approach among the co-investigators likely introduced an element of user-dependency. An additional important limitation of this study is the sample size. Machine learning experiments in image classification generally gain diagnostic accuracy when trained with very large data sets (e.g., subjects numbering in the tens of thousands), however large-scale advanced imaging MRI data sets are not readily available for such experiments. The need for large data sets is particularly relevant when applying deep learning approaches, such as convolutional neural networks. The decision by the authors to instead implement SVC and MLP models was an attempt to maximize the accuracy of the trained model in the setting of this limited training data set while lowering the likelihood of overfitting that may have occurred with a deep learning approach. Although some studies suggest that SVCs may outperform neural networks for image classification when utilizing relatively small datasets (43), in this study the SVC models achieved lower accuracies than the MLP models in the multiclass and head to head diagnostic challenges.
In summary, our trained multiclass MLP using VpNET2 can differentiate glioblastoma, brain metastasis, and CNS lymphoma with diagnostic accuracy approaching that of a neuroradiologist and provide approximately 19% increase in diagnostic yield when used as a CADx tool. Further study with larger data sets is required to improve diagnostic accuracy and demonstrate generalizability. Organized efforts by the radiology machine learning community to facilitate the sharing of anonymized diagnosis-specific, multimodal radiologic imaging in a HIPAA-compliant manner are needed to nurture this field of research.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: Institutional Review Board approval (ID #IF2169016) was obtained prior to this retrospective study.
References
- Sherwood PR, Stommel M, Murman DL, et al. Primary malignant brain tumor incidence and Medicaid enrollment. Neurology 2004;62:1788-93. [Crossref] [PubMed]
- Ranjan T, Abrey LE. Current management of metastatic brain disease. Neurotherapeutics 2009;6:598-603. [Crossref] [PubMed]
- Villano JL, Koshy M, Shaikh H, et al. Age, gender, and racial differences in incidence and survival in primary CNS lymphoma. Br J Cancer 2011;105:1414-8. [Crossref] [PubMed]
- Bernstein SH, Unger JM, Leblanc M, et al. Natural history of CNS relapse in patients with aggressive non-Hodgkin's lymphoma: a 20-year follow-up analysis of SWOG 8516 -- the Southwest Oncology Group. J Clin Oncol 2009;27:114-9. [Crossref] [PubMed]
- Mukundan S, Holder C, Olson JJ. Neuroradiological assessment of newly diagnosed glioblastoma. J Neurooncol 2008;89:259-69. [Crossref] [PubMed]
- Cha S. Neuroimaging in neuro-oncology. Neurotherapeutics 2009;6:465-77. [Crossref] [PubMed]
- Salcman M. Surgical resection of malignant brain tumors: who benefits? Oncology (Williston Park) 1988;2:47-56, 59-60, 63. [PubMed]
- Ferreri AJ, Reni M, Villa E. Therapeutic management of primary central nervous system lymphoma: lessons from prospective trials. Ann Oncol 2000;11:927-37. [Crossref] [PubMed]
- Yamasaki F, Kurisu K, Satoh K, et al. Apparent diffusion coefficient of human brain tumors at MR imaging. Radiology 2005;235:985-91. [Crossref] [PubMed]
- Guo AC, Cummings TJ, Dash RC, et al. Lymphomas and high-grade astrocytomas: comparison of water diffusibility and histologic characteristics. Radiology 2002;224:177-83. [Crossref] [PubMed]
- Hakyemez B, Erdogan C, Yildirim N, et al. Glioblastoma multiforme with atypical diffusion-weighted MR findings. Br J Radiol 2005;78:989-92. [Crossref] [PubMed]
- Toh CH, Chen YL, Hsieh TC, et al. Glioblastoma multiforme with diffusion-weighted magnetic resonance imaging characteristics mimicking primary brain lymphoma. Case report. J Neurosurg 2006;105:132-5. [Crossref] [PubMed]
- Hakyemez B, Erdogan C, Bolca N, et al. Evaluation of different cerebral mass lesions by perfusion-weighted MR imaging. J Magn Reson Imaging 2006;24:817-24. [Crossref] [PubMed]
- Weber MA, Zoubaa S, Schlieter M, et al. Diagnostic performance of spectroscopic and perfusion MRI for distinction of brain tumors. Neurology 2006;66:1899-906. Erratum in: Neurology 2006;67:920. [Crossref] [PubMed]
- Roberts HC, Roberts TP, Brasch RC, et al. Quantitative measurement of microvascular permeability in human brain tumors achieved using dynamic contrast-enhanced MR imaging: correlation with histologic grade. AJNR Am J Neuroradiol 2000;21:891-9. [PubMed]
- Jain R. Measurements of tumor vascular leakiness using DCE in brain tumors: clinical applications. NMR Biomed 2013;26:1042-9. [Crossref] [PubMed]
- Zacharaki EI, Wang S, Chawla S, et al. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 2009;62:1609-18. [Crossref] [PubMed]
- Zacharaki EI, Kanas VG, Davatzikos C. Investigating machine learning techniques for MRI-based classification of brain neoplasms. Int J Comput Assist Radiol Surg 2011;6:821-8. [Crossref] [PubMed]
- Tsolaki E, Svolos P, Kousi E, et al. Automated differentiation of glioblastomas from intracranial metastases using 3T MR spectroscopic and perfusion data. Int J Comput Assist Radiol Surg 2013;8:751-61. [Crossref] [PubMed]
- Sachdeva J, Kumar V, Gupta I, et al. Segmentation, feature extraction, and multiclass brain tumor classification. J Digit Imaging 2013;26:1141-50. [Crossref] [PubMed]
- Svolos P, Tsolaki E, Kapsalaki E, et al. Investigating brain tumor differentiation with diffusion and perfusion metrics at 3T MRI using pattern recognition techniques. Magn Reson Imaging 2013;31:1567-77. [Crossref] [PubMed]
- Alcaide-Leon P, Dufort P, Geraldo AF, et al. Differentiation of Enhancing Glioma and Primary Central Nervous System Lymphoma by Texture-Based Machine Learning. AJNR Am J Neuroradiol 2017;38:1145-50. [Crossref] [PubMed]
- Yamashita K, Yoshiura T, Arimura H, et al. Performance evaluation of radiologists with artificial neural network for differential diagnosis of intra-axial cerebral tumors on MR images. AJNR Am J Neuroradiol 2008;29:1153-8. [Crossref] [PubMed]
- Sachdeva J, Kumar V, Gupta I, et al. A dual neural network ensemble approach for multiclass brain tumor classification. Int J Numer Method Biomed Eng 2012;28:1107-20. [Crossref] [PubMed]
- El-Dahshan ESA, Mohsen HM, Revett K, et al. Computer-aided diagnosis of human brain tumor through MRI: A survey and a new algorithm. Expert Syst Appl 2014;41:5526-45. [Crossref]
- Cheng HL, Wright GA. Rapid high-resolution T(1) mapping by variable flip angles: accurate and precise measurements in the presence of radiofrequency field inhomogeneity. Magn Reson Med 2006;55:566-74. [Crossref] [PubMed]
- Patlak CS, Blasberg RG. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. Generalizations. J Cereb Blood Flow Metab 1985;5:584-90. [Crossref] [PubMed]
- Wu O, Østergaard L, Weisskoff RM, et al. Tracer arrival timing-insensitive technique for estimating flow in MR perfusion-weighted imaging using singular value decomposition with a block-circulant deconvolution matrix. Magn Reson Med 2003;50:164-74. [Crossref] [PubMed]
- Woolrich MW, Jbabdi S, Patenaude B, et al. Bayesian analysis of neuroimaging data in FSL. Neuroimage 2009;45:S173-86. [Crossref] [PubMed]
- Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011;12:2825-30.
- Kohli M, Prevedello LM, Filice RW, et al. Implementing Machine Learning in Radiology Practice and Research. AJR Am J Roentgenol 2017;208:754-60. [Crossref] [PubMed]
- Gaddikeri S, Gaddikeri RS, Tailor T, et al. Dynamic Contrast-Enhanced MR Imaging in Head and Neck Cancer: Techniques and Clinical Applications. AJNR Am J Neuroradiol 2016;37:588-95. [Crossref] [PubMed]
- Iannotti F, Fieschi C, Alfano B, et al. Simplified, noninvasive PET measurement of blood-brain barrier permeability. J Comput Assist Tomogr 1987;11:390-7. [Crossref] [PubMed]
- Bazyar S, Ramalho J, Eldeniz C, et al. Comparison of Cerebral Blood Volume and Plasma Volume in Untreated Intracranial Tumors. PLoS One 2016;11:e0161807. [Crossref] [PubMed]
- Server A, Orheim TE, Graff BA, et al. Diagnostic examination performance by using microvascular leakage, cerebral blood volume, and blood flow derived from 3-T dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging in the differentiation of glioblastoma multiforme and brain metastasis. Neuroradiology 2011;53:319-30. [Crossref] [PubMed]
- Abe T, Mizobuchi Y, Nakajima K, et al. Diagnosis of brain tumors using dynamic contrast-enhanced perfusion imaging with a short acquisition time. Springerplus 2015;4:88. [Crossref] [PubMed]
- Bhujwalla ZM, Artemov D, Glockner J. Tumor angiogenesis, vascularization, and contrast-enhanced magnetic resonance imaging. Top Magn Reson Imaging 1999;10:92-103. [Crossref] [PubMed]
- Liao W, Liu Y, Wang X, et al. Differentiation of primary central nervous system lymphoma and high-grade glioma with dynamic susceptibility contrast-enhanced perfusion magnetic resonance imaging. Acta Radiol 2009;50:217-25. [Crossref] [PubMed]
- Toh CH, Wei KC, Chang CN, et al. Differentiation of primary central nervous system lymphomas and glioblastomas: comparisons of diagnostic performance of dynamic susceptibility contrast-enhanced perfusion MR imaging without and with contrast-leakage correction. AJNR Am J Neuroradiol 2013;34:1145-9. [Crossref] [PubMed]
- Law M, Cha S, Knopp EA, et al. High-grade gliomas and solitary metastases: differentiation by using perfusion and proton spectroscopic MR imaging. Radiology 2002;222:715-21. [Crossref] [PubMed]
- Cha S, Lupo JM, Chen MH, et al. Differentiation of glioblastoma multiforme and single brain metastasis by peak height and percentage of signal intensity recovery derived from dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. AJNR Am J Neuroradiol 2007;28:1078-84. [Crossref] [PubMed]
- Rodriguez Gutierrez D, Awwad A, Meijer L, et al. Metrics and textural features of MRI diffusion to improve classification of pediatric posterior fossa tumors. AJNR Am J Neuroradiol 2014;35:1009-15. [Crossref] [PubMed]
- Shao Y, Lunetta RS. Comparison of support vector machine, neural network, and CART algorithms for the land-cover classification using limited training data points. ISPRS J Photogramm Remote Sens 2012;70:78-87. [Crossref]


