Abstract
BACKGROUND AND PURPOSE: In multiple sclerosis, gadolinium enhancement is used to classify lesions as active. Regarding the need for a standardized and accurate method for detection of multiple sclerosis activity, we compared 2D-spin-echo with 3D-gradient-echo T1WI for the detection of gadolinium-enhancing MS lesions.
MATERIALS AND METHODS: Fifty-eight patients with MS were prospectively imaged at 3T by using both 2D-spin-echo and 3D-gradient recalled-echo T1WI in random order after the injection of gadolinium. Blinded and independent evaluation was performed by a junior and a senior reader to count gadolinium-enhancing lesions and to characterize their location, size, pattern of enhancement, and the relative contrast between enhancing lesions and the adjacent white matter. Finally, the SNR and relative contrast of gadolinium-enhancing lesions were computed for both sequences by using simulations.
RESULTS: Significantly more gadolinium-enhancing lesions were reported on 3D-gradient recalled-echo than on 2D-spin-echo (n = 59 versus n = 30 for the junior reader, P = .021; n = 77 versus n = 61 for the senior reader, P = .017). The difference between the 2 readers was significant on 2D-spin-echo (P = .044), for which images were less reproducible (κ = 0.51) than for 3D-gradient recalled-echo (κ = 0.65). Further comparisons showed that there were statistically more small lesions (<5 mm) on 3D-gradient recalled-echo than on 2D-spin-echo (P = .04), while other features were similar. Theoretic results from simulations predicted SNR and lesion contrast for 3D-gradient recalled-echo to be better than for 2D-spin-echo for visualization of small enhancing lesions and were, therefore, consistent with clinical observations.
CONCLUSIONS: At 3T, 3D-gradient recalled-echo provides a higher detection rate of gadolinium-enhancing lesions, especially those with smaller size, with a better reproducibility; this finding suggests using 3D-gradient recalled-echo to detect MS activity, with potential impact in initiation, monitoring, and optimization of therapy.
ABBREVIATIONS:
- FSPGR
- fast-spoiled gradient recalled
- GRE
- gradient recalled-echo
- SE
- spin-echo
MR imaging is widely used in multiple sclerosis and has become an established tool not only for diagnosis but also for disease monitoring.1 MR imaging–defined disease activity, in conjunction with clinical status, can be used to ensure treatment optimization by identifying high-risk patients or poor responders during follow-up,2,3 which is important given that MS is a chronic disease for which expensive treatments are now used. There is an urgent need for an accurate and standardized MR imaging methodology among centers to better characterize and follow intrapatient changes longitudinally in personalized medicine and to facilitate analysis of large standardized datasets, which could help define predictors of disease evolution and long-term effects of therapies. Several attempts to improve and standardize MR imaging protocols have been published,4⇓–6 and several national cohorts are being developed (MAGNetic resonance Imaging in Multiple Sclerosis centers in Europe,7 L'Observatoire Français de la Sclérose en Plaques in France8), all sharing the objective of creating a central library documented by using high-quality homogeneous MR imaging examinations with demographic and clinical evaluations.
Despite published imaging recommendations,4⇓–6 recent technical developments suggest a careful re-examination of imaging methods and parameters. One significant development is the successful implementation of single-slab 3D sequences,9 which have become more commonplace with stronger field strengths (3T), better gradients, and improved receiver coil arrays. For example, the single-slab version of the 3D-T2-FLAIR has been shown to be better than conventional 2D-T2-FLAIR in terms of contrast-to-noise ratio, lesion detection, and homogeneity of CSF suppression.10⇓–12 Due to its higher sensitivity for lesion detection, 3D-T2-FLAIR is increasingly being used in MS and it also improves comparison across time points through easier registration of images to be displayed within the same geometric frame.6
While 3D-T1-weighted imaging sequences such as 3D fast-spoiled gradient recalled (FSPGR) and magnetization-prepared rapid acquisition of gradient echo have been in use for a long time and are widely used in clinical brain imaging,13 they are not routinely used in MS for visualization of active lesions after gadolinium injection. This practice is likely a result of theoretic14 and clinical studies15⇓–17 that demonstrated that 3D gradient recalled-echo (GRE) sequences could miss contrast enhancement compared with “conventional” 2D spin-echo (SE) images. While it is true that there are fundamental differences in the contrast behavior of the GRE and SE techniques,14 the dogma favoring 2D-SE over 3D-GRE for contrast enhancement of MS lesions comes from early work by using an older generation of imaging hardware and sequences.15⇓⇓–18 The increased availability of higher SNR 3T imaging systems and the improved contrast effect of gadolinium agents at 3T compared with 1.5T19⇓⇓–22 suggest a reconsideration of 2D-versus-3D imaging. High-performance imaging hardware and developments in parallel imaging permit whole-brain coverage with thin-section thickness by using 3D-GRE sequences in relatively short scanning times. The multiplanar reformatting capabilities with high SNR and contrast make 3D sequences appealing. In addition, Kakeda et al23 found significantly more metastases with contrast-enhanced 3D FSPGR than with 2D-SE imaging at 3T, especially small metastases of <3 mm in diameter. A direct comparison of 3D-GRE and 2D-SE T1-weighted sequences to depict active MS lesions after gadolinium injection has not been conducted, to our knowledge.
Consequently, recognizing the need for a standardized optimal detection of MS activity among centers to better guide therapy, we aimed at comparing the detection of gadolinium-enhancing lesions between 3D-GRE and conventional 2D-SE sequences. To achieve this goal, we combined an experimental approach in patients with MS with theoretic models analyzing the contrast-enhancement performance of 3D-GRE versus 2D-SE.
Materials and Methods
Subjects
Fifty-eight patients with clinically definite MS (48 women and 10 men; mean age, 39.3 years; range, 18–78 years) were prospectively included between March and October 2012. All were scanned when they were admitted to our hospital either to monitor treatment efficacy (n = 34) or because of a recent relapse (n = 24). Fifty had relapsing-remitting MS, and 8 had secondary-progressive MS. Disease duration ranged from 1 month to 39 years (mean, 10 years), and the median Expanded Disability Status Scale score was 2 (range, 0–8). All patients were already under disease-modifying treatment before MR imaging: Twenty-nine patients were treated with natalizumab; 9, with fingolimod; and the remaining 20 received interferon. Corticosteroids were not administered within 1 month before MR imaging, and even patients presenting with new neurologic symptoms were scanned before steroid administration. Both the patients and their relatives were informed that the patient's data might be used in clinical research studies, and all gave informed consent for their data to be analyzed after anonymization.
MR Imaging Acquisition
Brain imaging was performed on a whole-body MR imaging system operating at 3T (Discovery MR750w; GE Healthcare, Milwaukee, Wisconsin) equipped with high-performance gradients (maximum slew rate of 200 mT/m/ms and maximum strength of 50 mT/m) and using a 32-channel phased array head coil. The scanning protocol included a T2-FLAIR sequence, a pregadolinium 3D T1, and 2 postgadolinium sequences referred to as 2D-SE and 3D-GRE. The gadolinium-containing agent (gadoterate meglumine, Dotarem; Guerbet, Aulnay-sous-Bois, France) was injected manually at 0.2 mL/kg of body weight, and a 5-minute delay was observed before the acquisition of the first postinjection sequence. Because the delay between contrast injection and image acquisition may influence enhancement,24 the order of the 2D-SE and 3D-GRE was randomly chosen, with half of subjects being scanned with 2D-SE first and the other half, with 3D-GRE first. The following imaging parameters were used after administration of Dotarem—2D-SE refers to an axial T1 spin-echo sequence: 38 sections; thickness = 3 mm; gap = 1 mm; FOV = 22 cm; matrix = 320 × 224; voxel size = 0.7 × 1 × 3 mm3; TR/TE = 400/6.8 ms; bandwidth = 50 KHz; echo-train length = 2; refocusing flip angle = 111°; NEX = 2; parallel imaging factor = 1.5; scan time = 2 minutes 16 seconds. The sequence was acquired with flow-compensation gradients. 3D-GRE refers to a single-slab axially acquired T1 FSPGR: 128 sections; thickness = 1.2 mm; no gap; FOV = 24 cm; matrix = 288 × 288; voxel size = 0.8 × 0.8 × 1.2 mm3; TR/TE = 7.8/3.2 ms; bandwidth = 31 KHz; refocusing flip angle = 10°; NEX = 1; parallel imaging factor = 2; scan time = 2 minutes 24 seconds.
These parameters were optimized and adjusted for routine clinical imaging, and the matched acquisition times allowed a fair comparison between the 2 sequences.
Image Analysis
2D-SE and 3D-GRE after gadolinium injection were analyzed randomly and in a blinded fashion, with a minimum interval of 10 days between the 2 readings to avoid any recall bias. The analysis was done by 2 readers, independently, blinded to any clinical information. One reader was a junior radiologist in residency training in radiology with 2 years' experience in MR imaging, including 6 months as a neuroradiology resident (A.C.), and the other was a senior neuroradiologist with >9 years' experience in MR imaging, especially in neuroimaging (T.T.). Subsequently, a final interpretation was made in consensus, with the addition of a third experienced neuroradiologist (V.D., with >25 years' experience) in case of disagreements.
The readers were asked to count the gadolinium-enhancing lesions and to measure their maximal diameter. The precontrast T2-FLAIR sequence was always available during the interpretation because an enhancing lesion had to be seen on T2 FLAIR and to appear with high signal intensity on postgadolinium T1WI, irrespective of its maximum length or the number of sections in which the lesion was visible. The enhancing lesions were categorized according to their anatomic location in infratentorial, periventricular, juxtacortical, and other deep white matter locations as defined previously.22 Finally, the pattern of enhancement was also categorized as uniform or ringlike.5 The 3D-GRE volume could be reformatted in arbitrary planes to assess the presence of lesions, a unique advantage of 3D sequences.
In the subgroup of lesions of >5 mm in diameter that were found to enhance on both the 2D-SE and the 3D-GRE, identical ROIs were placed by 1 reader within the lesion and the adjacent normal-appearing white matter, to compute relative contrast defined as (Signallesion − SignalWM)/(Signallesion + SignalWM). This metric was preferred rather than the contrast-to-noise ratio because noise cannot be accurately measured when parallel imaging is used without duplicating the sequence to measure the noise on the image difference.25
Simulations
To understand the theoretic signal and contrast behavior of the sequences used above after gadolinium injection, we performed Matlab (MathWorks, Natick, Massachusetts) simulations of lesion SNR and lesion-WM relative contrast by implementing the signal equation for 2D-SE and 3D-GRE, similar to that of Mugler and Brookeman.13 We used a T1 value of 1300 ms and a T2 value of 100 ms to define a theoretic MS lesion precontrast at 3T.26 We used the data of Yu et al27 to set the relaxivity values of Dotarem at 3T as r1 = 5.5 mmol−1s−1 and r2 = 6.25 mmol−1s−1 and varied the gadolinium concentration values from 0 to 5 mmol/L. We simulated 2 scenarios: 1) The lesion is large and occupies the entire voxel in both the 2D and 3D sequences, and 2) the lesion is small and occupies the entire voxel in 3D but not in 2D sequences (ie, 2D lesion volume < 2D voxel volume and 3D lesion volume = 3D voxel volume). The former is the worst-case scenario, and the latter is more realistic for cases we encountered and the scan parameters we used. Note that the 3D and 2D voxel sizes on the acquired images and used in these simulations were 0.8 × 0.8 × 1.2 mm3 and 0.7 × 1.0 × 3.0 mm3, with 3D voxel volume being ∼2.5 times smaller than the 2D voxel volume.
Statistical Analysis
Nonparametric tests were used because the number of lesions did not follow a Gaussian distribution. Statistical differences in lesion counting were compared patient-wise by using the Wilcoxon signed ranked test for matched pairs. Differences were first tested between 3D-GRE and 2D-SE for the junior and the senior radiologists and then between readers for the 3D-GRE and 2D-SE by using Bonferroni corrections to adjust for multiple (4 pair-wise) comparisons. The interreader agreement between the junior and the senior readers with respect to the number of gadolinium-enhancing lesions detected on 3D-GRE and 2D-SE was assessed by calculating the Cohen κ test. Finally, differences in the proportions of gadolinium-enhancing lesions detected with 3D-GRE versus 2D-SE in terms of location, pattern of enhancement, and lesion size were compared with the McNemar test for paired data, while relative contrast values of gadolinium-enhancing lesions detected with 3D-GRE versus 2D-SE were compared with the Wilcoxon signed ranked test. Statistical analyses were performed by using R statistical computing software, Version 3.0.0 (http://www.r-project.org). A corrected P value ≤ .05 was statistically significant.
Results
In Vivo Study in Patients
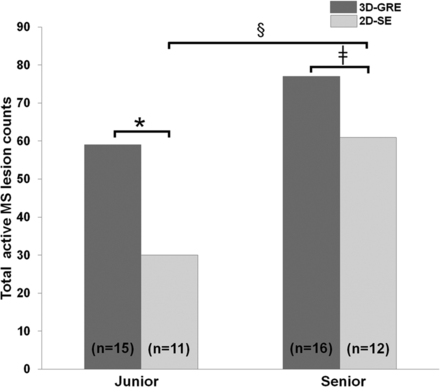
Both the junior and the senior readers consistently reported more gadolinium-enhancing lesions on 3D-GRE compared with 2D-SE (Fig 1). The junior radiologist reported 59 enhancing lesions in 15 patients on 3D-GRE, which was 96% more compared with 2D-SE (30 lesions in 11 patients), while the senior radiologist reported 77 enhancing lesions in 16 patients on 3D-GRE, which was 26% more compared with 2D-SE (61 lesions in 12 patients). For the junior and the senior readers, 4 patients would have been missed as “active” when using only 2D-SE. Patient-wise analysis showed that these differences were statistically significant, with a higher number of patients with more enhancing lesions on 3D-GRE compared with 2D-SE both for the junior and senior radiologists (P = .021 and P = .017, respectively). All lesions counted on 2D-SE were also seen on 3D-GRE for both readers; thus, 29 and 16 lesions for the junior and the senior readers, respectively, were supplementary lesions reported on 3D-GRE only.
Total count of gadolinium-enhancing lesions on 3D-GRE (dark gray) and 2D-SE (light gray) is plotted for the junior and the senior readers. The data in brackets are the number of patients with at least 1 gadolinium-enhancing lesion detected on 3D-GRE (dark gray) or 2D-SE (light gray). The asterisk, double dagger, and section sign indicate corrected P values of .021, .017, and .044, respectively, obtained by the Wilcoxon test for matched pairs.
The senior reader had a significantly better lesion-detection rate compared with the junior reader on 2D-SE (P = .044), while the difference did not reach statistical significance on 3D-GRE (P = .078). In line with this observation, the agreement between the 2 readers was better for the 3D-GRE (κ = 0.65, indicating good agreement; 95% confidence interval, 0.50 < intraclass correlation coefficient < 0.79) than for the 2D-SE (κ = 0.51, indicating fair agreement; 95% confidence interval, 0.36 < intraclass correlation coefficient < 0.64).
To determine which factors could contribute to the higher rate of gadolinium-enhancing lesions detected on the 3D-GRE compared with the 2D-SE, we compared the features of gadolinium-enhancing lesions detected by 3D-GRE and 2D-SE for qualitative and quantitative differences by using the final consensual interpretation.
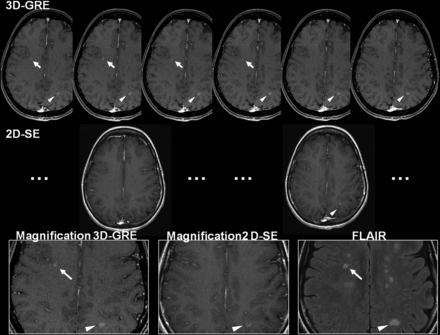
Qualitatively, the additional gadolinium-enhancing lesions seen on 3D-GRE were sometimes totally invisible on 2D-SE (n = 11; Fig 2 for an illustrative example) and sometimes were not scored on the 2D-SE in the first reading while retrospectively detected during the final consensual side-by-side comparison (n = 5). Such misdiagnoses on 2D-SE were attributed to difficulty associated with pulsation artifacts in the venous sinus that were less dominant on 3D-GRE (Fig 3) or to confusion with enhancing cortical vessels on 2D-SE. Vessel distinction was not problematic on 3D-GRE due to decreased partial volume effects and 3D reformats that allowed scrolling through images in 3D to distinguish linear vascular structures present in multiple planes from enhancing MS lesions.
Illustrative example of a small right juxtacortical lesion (arrow) that was scored as gadolinium-enhancing by the 2 readers on 3D-GRE, being visible on 4 consecutive sections, while it was fully invisible even retrospectively on 2D-SE. The contralateral larger lesion (arrowhead) was counted as enhancing on both 3D-GRE and the 2D-SE. In this case, 2D-SE was acquired before 3D-GRE.
Illustrative example of a right cerebellar lesion (arrow) that was scored as gadolinium-enhancing by the 2 readers on the 3D-GRE but not on 2D-SE during the independent blinded reading sessions. Retrospectively, subtle gadolinium enhancement can also be seen on 2D-SE but in a single section (as opposed to 5 sections on 3D-GRE), where it could easily be misdiagnosed as ghosting artifacts induced by the adjacent sinus. The contralateral lesion (arrowhead) was counted as gadolinium-enhancing on both 3D-GRE and the 2D-SE. In this case, 2D-SE was acquired before 3D-GRE.
Quantitatively, 3D-GRE showed significantly more small lesions (<5 mm) than 2D-SE in patient-wise analysis (P = .04), while there was no difference for larger lesions (>5 mm). All the other features were similar for gadolinium-enhancing lesions seen on 2D-SE or 3D-GRE. Specifically, there was no confounding effect induced by the delay of acquisition after the injection24 because exactly 50% of patients with enhancing lesions were scanned with 3D-GRE first and the other 50%, with 2D-SE first. There was a trend, though not significant, for a higher detection on 3D-GRE, even in the subgroup explored with 3D first (n = 46 versus n = 31). Furthermore, the distribution of gadolinium-enhancing lesions was identical between 3D-GRE and 2D-SE in terms of location (for 3D-GRE and 2D-SE, respectively: 20.8% and 21.3% in periventricular; 39% and 42.6% in deep WM; 28.6% and 27.8% in juxtacortical; and 11.6% and 8.3% in infratentorial locations) and pattern of enhancement (for 3D-GRE and 2D-SE, respectively: 56% and 47% with homogeneous enhancement; 44% and 53% with ringlike enhancement). Finally, measurement of the gadolinium-enhancing lesions large enough to avoid any difficulty with region-of-interest placement (diameter of >5 mm) and seen with both 3D-GRE and 2D-SE (n = 15 lesions) showed similar relative contrast between gadolinium-enhancing lesions and the adjacent white matter (contrast = 0.16 ± 0.08 on 2D-SE versus 0.15 ± 0.07 on 3D-GRE, P = not significant).
Simulations and Theoretic Models
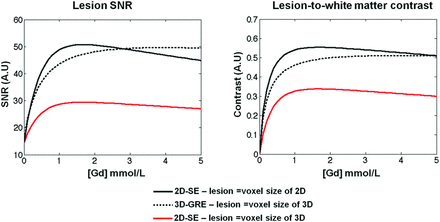
Figure 4 summarizes the results of theoretic simulations of SNR and contrast behavior for the 2D and 3D sequences by using the scan parameters in the actual imaging experiments. For a simulated MS lesion occupying the whole voxel on 2D-SE and 3D-GRE, the lesion-SNR and the lesion-to–white matter contrast as a function of the gadolinium concentration within this lesion were close. The 2D-SE still provided slightly better performance for lesions accumulating a small amount of gadolinium (low gadolinium concentration), but even in this situation, there was not a major drop in SNR and contrast on 3D-GRE, despite a voxel size >2.5 times smaller than that on 2D-SE. If we consider small lesions that occupy the entire 3D-GRE voxel but only a portion of the 2D-SE voxel, the simulations predict a significant decrease in lesion SNR and lesion-to–white matter contrast on 2D-SE (solid red), which was roughly in agreement with our experimental results. While the true gadolinium concentration in vivo in an MS lesion is unknown, some authors have estimated that the typical gadolinium dose of 0.2 mL/Kg of body weight leads to a target tissue concentration of ≥0.5 mmol/L.28 At this gadolinium concentration, the models predicted that the SNR of a large lesion on 3D-GRE was decreased by 9.5% compared with 2D-SE but was increased by 46% for a small lesion. Similarly, lesion-WM contrast on 3D-GRE was decreased by 16% compared with 2D-SE for a large lesion but was increased by 45% for a small lesion.
Plots of lesion SNR and lesion-to–white matter relative contrast (same definition as that for in vivo experiments (ie, [Slesion − SWM]/[Slesion + SWM]) as a function of the concentration of gadolinium within theoretic MS lesions assumed to fulfill a whole-voxel size in 2D-SE (dark solid line) and 3D-GRE (dark dotted line). If a lesion the size of the 3D voxel is simulated, the dark dotted line still shows the lesion SNR and lesion-to–white matter contrast on 3D-GRE but the 2D-SE shows a far lower signal and contrast (red solid line).
Discussion
We found that the 3D-GRE sequence after injection of gadolinium in patients with MS detected more enhancing lesions and more patients with enhancing lesions than the standard 2D-SE sequence, which failed to detect some active lesions. This result is in contrast to earlier ones showing better sensitivity of the 2D SE sequence to contrast enhancement15,17 and suggests the use of a 3D-GRE-type sequence after gadolinium injection for imaging patients with MS to improve the quality and consistency of routine scanning.
Currently, gadolinium has a pivotal role in MS to assess the radiologic disease activity,1 which is used in turn for diagnosis,29 guiding therapeutic strategies,2,3,30 and as a surrogate marker to evaluate treatment efficacy in clinical trials.31 After a first clinical event suggestive of MS (clinically isolated syndrome), detection of a single gadolinium-enhancing lesion can argue for “dissemination in time,”32 which could allow fulfilling diagnostic criteria of MS, which, in turn, could accelerate early therapy. At later stages, a single gadolinium-enhancing lesion can provide a strong argument to optimize therapy by identifying an aggressive course or a suboptimal treatment response at follow-up, both requiring escalation of therapy.3,30 Any additional sign of activity depicted with 3D-GRE could therefore have a strong impact in clinical practice. Whether the additional sensitivity that we are reporting here translates to such clinical relevance remains a crucial question. The current study could be viewed as the first step to be followed up with larger studies on homogeneous populations to ascertain whether the 3D-GRE could really lead to an earlier diagnosis or have any therapeutic consequences.
While earlier studies reported that contrast enhancement might be less conspicuous on GRE images compared with SE,15⇓–17 those studies were performed on older scanners. We revisited this possibility, keeping in mind the improvements in scanner hardware and pulse sequences. The higher field strength at 3T affects the relaxation time of both enhanced and nonenhanced tissues, but disproportionately, with a greater relative T1 shortening effect after gadolinium at 3T than at lower field strengths.21 The linear increase of signal-to-noise ratio with the field strength is another factor that might improve the quality of higher resolution images such as 3D-GRE and, in turn, the conspicuity of small lesions. The higher field strength also enhances parallel imaging performance by reducing noise enhancement in reconstruction,33 which can be further improved by the 32-channel coil. Altogether, the fundamental differences between the contrast behaviors of the SE and GRE techniques reported earlier14 might not necessarily result in the inferiority of GRE, and the higher spatial resolution of 3D-GRE could even overtake SE for active lesion detection. In our study, the thinner section thickness and reduced partial volume averaging seem to be the main factors associated with 3D-GRE performance regarding the identical experimental values of relative contrast but the significantly higher rate of small lesions of <5 mm on 3D-GRE. Some of the lesions were completely invisible on 2D-SE, even after retrospective reading. In our experimental design, the readers could not be blinded to the sequence type (2D-SE versus 3D-GRE) because it is easily recognizable, and that factor could have potentially biased the results of the reading. Nevertheless, the simulation results, which are consistent with the in vivo data, strengthen our experimental findings. From these simulations, in line with the literature,14 2D-SE is expected to provide slightly higher SNR and contrast if the amount of gadolinium is low and the lesion occupies the whole voxel. Nevertheless, no lesions seen on 2D-SE were missed by 3D-GRE. With the parameters of the sequences routinely used in clinical imaging at 3T and assuming a different lesion/volume contribution, which is the case for small lesions fully captured on the 3D voxel but only partly on the 2D voxel, the simulations predict that the SNR and the contrast of an enhancing lesion will be significantly better by using 3D-GRE than 2D-SE.
Moving to 3D datasets for postgadolinium imaging in patients with MS could offer other advantages in addition to its higher sensitivity to enhancing lesions. Such images seem easier to read on the basis of our junior and senior readers' results, with probably less training required to distinguish noise, artifacts, anatomic variants, and true lesions. For example, the pulsatile flow artifacts seen on 2D-SE, even if not fully absent, were reduced on 3D-GRE because of the shorter TEs used in 3D-GRE. Furthermore, with 3D being more amenable to image registration, the 3D images could be easily displayed in a fixed-registration framework, facilitating longitudinal follow-up and standardization. Finally, because noncontrast 3D-GRE is becoming more frequently used in MS to monitor atrophy,6 it makes sense to use the same sequence after contrast injection for an accurate pre/post comparison.
This study has limitations from the limited number of active lesions, which nevertheless corresponds to the classic recruitment of patients with MS, who are more often controlled by therapy. Such a study also has an intrinsic limitation from the absence of a valid reference standard (histopathologic confirmation) to define which lesions are truly active or inactive. Therefore, we did not compute true sensitivity and specificity, but we compared absolute numbers of lesions detected with each sequence, considering each lesion is truly active, because misdiagnoses can be reasonably excluded during the final consensual interpretation with expert readers. Furthermore, the theoretic approach provides additional evidence to support our experimental findings, even in the absence of a valid criterion standard. Last, we did not use the recently developed class of 3D-TSE sequences, which could combine the advantage of the SE and the 3D acquisitions while using variable flip angles to optimize contrast and reduce the specific absorption rate.34,35 While these sequences are promising,34 they are still less widely available compared with the 3D-GRE we used here, availability being important for standardization of MS imaging protocols among a large number of centers with different vendors. These 3D-TSE sequences are also longer than the 3D-GRE-based T1 and may have more motion artifacts due to subject motion and blurring due to the long echo-trains.36 3D-GRE T1 imaging is thus an easy start for improving the MS protocol for clinical routine or research purposes after gadolinium injection.
Conclusions
In summary, we have demonstrated that the contrast-enhanced 3D T1 FSPGR sequence can detect significantly more active demyelinating lesions than the conventional axial T1 SE sequence at 3T in patients with MS. We suggest reconsidering a 3D sequence post-gadolinium injection, which can be easily standardized among centers, instead of 2D-SE, to improve routine scanning for personalized medicine and also clinical research studies.
Acknowledgments
We thank Alice Doreille for her implication in this MS imaging study.
Footnotes
Disclosures: Monojkumar Saranathan—UNRELATED: Other: research support from GE Healthcare.* Aurelie Ruet—UNRELATED: Board Membership: Biogen Idec; Consultancy: Teva; Travel/Accommodations/Meeting Expenses Unrelated to Activities Listed: Biogen Idec, Novartis; OTHER RELATIONSHIPS: I am a recipient of a European Committee for Treatment and Research In Multiple Sclerosis fellowship research grant. Bruno Brochet—UNRELATED: Board Membership: Biogen Idec, Teva; Grants/Grants Pending: Teva, Merck Serono, Bayer, Caridian; Payment for Lectures (including service on Speakers Bureaus): Novartis, Teva; Payment for Development of Educational Presentations: Novartis, Genzyme; Travel/Accommodations/Meeting Expenses Unrelated to Activities Listed: Novartis, Teva. *Money paid to the institution.
This work was supported by the FLI (ANR-11-INBS-0006) and the French Agence Nationale de la Recherche within the context of the Investments for the Future Program, referenced ANR-10-LABX-57 and named TRAIL.
Paper previously presented in part at: Annual Meeting of the International Society for Magnetic Resonance in Medicine and European Society for Magnetic Resonance in Medicine and Biology (eposter 6954), May 10-16, 2014; Milan, Italy; and Annual Meeting of the French Society of MS (Association pour la Recherche contre la Sclérose en Plaques), May 23, 2014; Paris, France.
Indicates open access to non-subscribers at www.ajnr.org
REFERENCES
- Received June 25, 2014.
- Accepted after revision September 1, 2014.
- © 2015 by American Journal of Neuroradiology