Abstract
Objective
Brain tissue segmentation by conventional threshold-based techniques may have limited accuracy and repeatability in older subjects. We present a new multispectral magnetic resonance (MR) image analysis approach for segmenting normal and abnormal brain tissue, including white matter lesions (WMLs).
Methods
We modulated two 1.5T MR sequences in the red/green colour space and calculated the tissue volumes using minimum variance quantisation. We tested it on 14 subjects, mean age 73.3 ± 10 years, representing the full range of WMLs and atrophy. We compared the results of WML segmentation with those using FLAIR-derived thresholds, examined the effect of sampling location, WML amount and field inhomogeneities, and tested observer reliability and accuracy.
Results
FLAIR-derived thresholds were significantly affected by the location used to derive the threshold (P = 0.0004) and by WML volume (P = 0.0003), and had higher intra-rater variability than the multispectral technique (mean difference ± SD: 759 ± 733 versus 69 ± 326 voxels respectively). The multispectral technique misclassified 16 times fewer WMLs.
Conclusion
Initial testing suggests that the multispectral technique is highly reproducible and accurate with the potential to be applied to routinely collected clinical MRI data.
Similar content being viewed by others
Introduction
White matter lesions (WMLs) are commonly observed on MR images of older people. Their association with declining cognitive ability, future stroke risk and dementia is well recognised. WMLs can be assessed using subjective rating scales, but the wider availability of more detailed MR brain imaging has led to increasing attempts to measure WML volumes.
Many studies measure WML volume using a single MR sequence [1–6]. Some delineate WMLs using a region-growing algorithm based on pixel intensity in FLAIR images, followed either by manual editing [1, 6, 7] or by mixture modelling–based algorithms [4]. Other methods use local thresholds in each slice to mark the lesions and borders [5]. While these techniques are typically methodologically simple, the resulting segmentations are prone to operator bias, have low precision and are time consuming [8].
Combining sequences may increase the reliability of tissue segmentation and reduce the reliance on observer input. Such approaches include probabilistic methods [9], neural networks [10], genetic algorithms [11], fuzzy segmentation [12], Parzen windows [13], the Markov field random model, and the expectation-maximisation algorithm [14]. These methods demand complex computational efforts to achieve good reproducibility and publications often fail to mention practicalities such as processing time or the amount of manual editing required. Finally, it is unclear if atrophy and focal lesions, which are common in older subjects and may interfere with the image processing algorithms, affect the accuracy of these techniques.
We developed a completely new approach to overcome these difficulties for a study of ageing [15]. We fused pairs of different MR sequences and modulated them in the red-green (RG) colour space thereby enhancing tissue discrimination. We compared this with WML segmentation performed by local thresholding of a FLAIR sequence, a commonly used approach.
Materials and methods
Subjects
We selected MR data sets from two previous studies: one of normal male volunteers aged 65–70 years and one of patients with mild stroke aged 50–90 years [16, 17]. The MR images were rated for WMLs using validated qualitative rating scales [18] by two experienced raters (JMW, KJF), blinded to each other [19]. Using these ratings, we randomly selected 14 subjects that represented the full range of WMLs and brain atrophy: eight from the normal male volunteers aged 65–70 years [20], and six from the patients with mild non-disabling stroke aged 50–90. [17] The average age of the 14 subjects was 73.3 ± 10 years. Both studies had Ethics Committee approval, and all subjects gave informed consent.
MRI
All MR images were obtained on a 1.5T GE Echospeed MRI system. These included sagittal T1-weighted (T1W), axial T2-weighted (T2W), T2*-weighted (T2*W) and FLAIR images (sequence details given in Table 1). The DICOM files were converted to data ANALYSE format. [21] The images from the stroke study were linearly interpolated from matrix 256 × 192 to 256 × 256.
Definition of WMLs
We identified WMLs as diffuse patchy areas in the white matter (WM), basal ganglia or brain stem, larger than 3 mm in diameter and hyperintense with respect to normal white and grey matter on T2W and FLAIR images [18].
Threshold-based WML segmentation on FLAIR
FLAIR-based thresholding relies on the change in tissue intensity at the WML boundary. To determine the best and most subject-specific thresholds, we sampled signal intensity in normal-appearing white matter using Analyze 8.1 software [21]. We placed two regions of interest (ROIs), each 5 × 5 mm square, bilaterally, in normal-appearing frontal white matter on all slices on which frontal white matter was visible, from the inferior frontal lobes to near the vertex. We grouped the slices into 12 adjacent pairs and calculated the mean ROI intensity for each slice pair. The subject’s WML threshold was set as six standard deviations (SDs) above the mean intensity, a method previously tested in our lab. This produced 12 thresholds per subject, from inferior frontal to near vertex.
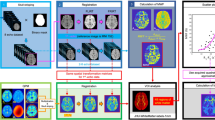
Central axial slice of a subject displaying the registration and fusion processes for MCMxxxVI. First row, left to right: T2W, T1W images and their fusion and modulation in red and green channels respectively; second row, left to right: T2*W and FLAIR images and their fusion and modulation in red and green channels
We produced three WML masks per patient using the maximum, minimum and mean of these 12 thresholds. To identify WMLs, we placed an auto-trace seed in normal white matter on a reference image and highlighted automatically all areas of intensity higher than the calculated threshold [3]. The WML edges were then automatically delineated by a region-growing algorithm from seeds placed manually on the lesions and set at the intensity range: min = calculated threshold, max = maximum intensity. We performed minimal manual editing to remove erroneously included areas (e.g. normal caudate nucleus) and repeated the above process without excluding any erroneous areas to test the influence of observer intervention. We also examined whether the FLAIR-derived thresholds were influenced by slice position by comparing the threshold values of the 12 slice pairs.
Multispectral data fusion and minimum variance quantisation (MVQ) technique
To develop our new technique, we selected two MR sequences that produced different intensity ranges in the tissues to be segmented and optimised their grey scale intensity range using Analyze 8.1 [21]. By trial and error, we found that the coloured RG combination of T2*W and FLAIR was good for differentiating WMLs and CSF and the combination of T1W and T2W was good for recognising CSF and white matter (Fig. 1). We registered the images using the 3-D Surface Registration module in Analyze 8.1, a surface-matching algorithm that is fast and robust, even in the presence of image noise [21]. We used T2W images as ‘base’ objects and ‘matched’ all other images (T1W, T2*W, FLAIR) to it. We transformed the sequence pairs to the red-green (RG) colour space [22], putting T2*W through the red channel and FLAIR through the green channel so that the resulting coloured volume contained the information of the fused T2*W and FLAIR volumes. We performed a similar process for the T1W and T2W sequence pair. We used the RG colour space because the human eye cones are most sensitive to the frequencies in this range of the visible spectrum [23]. Wyszecki and Stiles [24] measured luminous efficiency using six different techniques and demonstrated that the maximum luminous efficiency of the human eye is reached at wavelengths of around 550 nm, remaining at 80% or above up to the maximum value in the interval from 525 nm (green) to 670 nm (dark red).
To extract the brain from the fused RG volumes for further processing, we used T2*W images, as this provides good differentiation between brain-CSF and inner skull table and extracted the brain using the Object Extraction Tool in Analyze 8.1 [21]. To segment and quantify the WML and tissue volumes from the RG-modulated image, we developed a software module using MATLAB (Fig. 2) [25]. The initial fused volume had 65,535 different levels in the RG space, so a minimum variance quantisation (MVQ) algorithm, which uses Floyd-Steinberg’s ‘error diffusion dither principle’ [26] implemented by the MATLAB function ‘rgb2ind’ [25] was used to reduce colour depth to a more manageable number of levels. Orchard and Bouman showed MVQ to be the optimal clustering method because more colour map entries are allocated to densely populated areas in the colour space, and fewer entries are allocated to infrequent colours [27]. They demonstrated that MVQ achieved a time complexity of Nlog2 M, where N is the number of pixels in the image and M the number of the desired clusters in which the coloured image is dithered. Experimentally we determined that reducing the 65,535 levels to 32 clusters provided good reproducibility of all tissue classes (see graph, Fig. 2).
We then selected the clusters corresponding to the tissue of interest, e.g. WMLs. Moving the cursor over the WMLs in the quantised images showed the corresponding RG coordinate values on the graph (Fig. 2). One, or at most a few, prototypical clusters per subject are sufficient to identify the maximum and minimum RG coordinates of the required tissue on each axis. The software then automatically constructs the tissue segmentation mask from the quantised volume. Minimal manual editing was required to remove minimal false positive WMLs. In this paper we compare the WML volumes before and after manual editing. The video (multimedia file, see the ESM) demonstrates the process of extracting CSF and WMLs.
We named this method “MCMxxxVI” which stands for multispectral colouring modulation and variance identification and also represents the number 1936, reflecting the Lothian Birth Cohort 1936 study for which it was developed.
Observer variation and statistical analysis
Two trained observers (MVH and KJF), independently and blinded to each other’s results, performed WML segmentation using the MCMxxxVI method and FLAIR-derived thresholds (comparisons summarised in Table 2). The FLAIR thresholds derived from the three most caudal slice pairs were discarded from further analyses as they were clearly different (Fig. 3). We calculated the minimum (threshold 1) and maximum (threshold 2) per patient and also a single threshold equal to the average of all 168 thresholds (9 slice pairs, 14 subjects). The single threshold was evaluated because some studies use only one threshold for segmenting WMLs in all patients [2]. The resulting three WML masks were visually assessed, blind to threshold value and to each other, for accuracy by an experienced neuroradiologist (JMW) who determined the number of false positive and partial and total false negative WMLs per brain region.
We expressed the WML volumes derived from each method in voxels and calculated the mean, maximum and minimum differences using Bland-Altman analyses [28]. The effect of slice location and WML load on FLAIR-derived thresholds was explored using a linear mixed model and expressed as means with 95% confidence intervals (CI). Separate simple linear regression analyses were used to test for the effect of observer variation of RG boundary values on WML volume. The results of the intra- and inter-observer reliability tests are expressed in voxels, not cubic millimetres because of the inter-slice gap (5 mm), i.e. we did not at this stage apply stereology to interpolate between slices. Finally, we evaluated the effect of motion artefact and magnetic field heterogeneities [29] on WML volume measurement [13, 30].
Results
FLAIR-derived WML thresholds were influenced by both slice position and WML volume. The FLAIR derived threshold decreased by 2.52 (3.90, 1.14) units between sequential adjacent slice pairs from brain stem to vertex (P = 0.0004, Fig. 3), and increased by 3.14 (95% CI 1.79, 4.48) units for every voxel increase in WML volume (P = 0.0003, Table 3). In comparison, WML volume had no effect on RG levels or on WML volume using the MCMxxxVI (Table 3). FLAIR-derived thresholds were also affected by image contrast levels, indeed the image of one subject differed very much from the others because the contrast was extremely low (data not shown). Image contrast had no effect on MCMxxxVI-derived volumes.
There was good intra-rater repeatability for WML volumes calculated using the same FLAIR-derived threshold each time (SD of ±734 voxels) and with MCMxxxVI (SD of ±326 voxels) but not when an individual new FLAIR-derived threshold was used each time to segment the WMLs (SD of ±9,924 voxels, Table 4). This lowest intra-rater variation for the MCMxxxVI method occurred even without manual correction to remove any false positives/negatives (Table 4). We did not measure the inter-rater variability of the FLAIR-derived threshold techniques, as its intra-rater measurement variability was so much higher than the inter-rater variability of the MCMxxxVI method.
Visual comparison of WMLs identified using FLAIR-derived thresholds and MCMxxxVI showed that the range (y axis) of errors was 16 times greater for FLAIR-derived thresholds than for MCMxxxVI, i.e. both false positives and negatives were very frequent with the FLAIR-derived thresholds and increased with increasing WMLs but were rare with MCMxxxVI (Fig. 4). WML segmentation errors differed with brain region in both methods (data not shown), but the MCMxxxVI approach misclassified few lesions except in the pons, where a few errors occurred due to CSF flow artefacts.
Discussion
We present a novel multispectral image processing method, MCMxxxVI, for segmenting normal brain tissue and WMLs. We have demonstrated that slice position in the brain, WML load and focal lesions affect the accuracy of WML volume determined using FLAIR-derived thresholds but have little influence on the MCMxxxVI method. In this test sample of 14 subjects, deliberately chosen to represent a wide range of WMLs and accompanying features of ageing such as atrophy, the MCMxxxVI had the best intra-rater reliability. In contrast to FLAIR-derived thresholds, MCMxxxVI was not affected by focal signal extremes, number of WMLs, bias field heterogeneities or movement artefacts. It was fast and easy to use. Further testing is now warranted in much large populations.
The MCMxxxVI method uses an area of the two dimensional RG space that combines information from two different MR sequences chosen to optimise differences in signal between tissue types, i.e. it enhances their radiological properties. In addition to segmenting WMLs, the multispectral technique also enables extraction of intracranial volume, whole brain, CSF, grey matter and normal white matter volumes using different combinations of MR sequences. We used T2* and FLAIR to segment CSF and WMLs and T2 and T1 to segment normal grey and white matter. An added advantage is that use of two or more images in combination reduces the effects of distortions and noise found in a single sequence. Thus a FLAIR image with movement artefact or bias field heterogeneity impairs measurement of WML volume using a threshold, potentially rendering that subject’s data unusable. However, as it is unlikely that all sequences would be badly affected by movement or bias field heterogeneity, combining the FLAIR with a less movement-affected or bias field-affected T2* in the MCMxxxVI method attenuates any adverse effects present in one sequence and means that the subject’s data can still be used.
The strengths of our study are that we have compared our method with a conventional WML volume measurement method and examined the sources of error and variation with computational as well as visual assessment. We used carefully chosen images representing a wide range of WMLs and other abnormalities from normal older subjects and patients with stroke. To our knowledge, no previous studies have assessed WML segmentation as comprehensively. The weaknesses include the small sample, the use of data from one scanner (albeit with different sequence details) and the prototypical stage of the MCMxxxVI method placing it at a disadvantage compared with more established methods such as FLAIR thresholding. We have not performed extensive testing of the influence of WML volume on RG space boundaries or methods to remove the minor misclassification of WMLs in the posterior fossa due to CSF flow artefacts. However these data now warrant testing in other larger cohorts and other refinements.
The problems that affect FLAIR-derived threshold WML volume measurement are likely to affect all threshold-based techniques. Thus, in noisy MRIs or in patients with severe WMLs, a suitable area of homogeneous, normal appearing white matter may be difficult to find and a single sequence to determine the threshold is very vulnerable to RF inhomogeneities [29]. Focal lesions that create marked focal variations in signal strength, such as infarcts, large volume ventricles and haemosiderin deposits in the basal ganglia, also affected FLAIR-derived thresholds and increase WML volume variability. The MCMxxxVI method reduced considerably the influence of RF inhomogeneities and noise, improving the signal to noise ratio, thus enhancing tissue differentiation. Genuine changes in normal-appearing white matter occur with increasing age, and possibly also with increasing WMLs independently of age, and are likely to affect all threshold-based WML volume measurement and tissue segmentation techniques. Thus in subjects with more WMLs, higher thresholds may lead to an underestimation of WML lesion volume and greater misclassification of WMLs, introducing systematic bias to studies of factors associated with WML volume.
We have demonstrated that the MCMxxxVI method is fast, accurate, avoids the problems caused by many of the biological features that affect semi-automated threshold-based segmentation of FLAIR images, and uses calculations that are relatively simple to apply. The use of two MR sequences reduces problems due to movement and field heterogeneity during scan acquisition, and the lengthy processing times and computational requirements of traditional methods. This work suggests that the multispectral colour fusion of two or more MR images is a powerful tool to be explored and considered further in tissue segmentation.
References
Gootjes L, Teipel SJ, Zebuhr Y, Schwarz R, Leisinger G, Scheltens P, Moller HJ, Hampel H (2007) Regional distribution of white matter hyperintensities in vascular dementia, Alzheimer’s disease and healthy aging. Dement Geriatr Cogn Disord 18:180–188
Hugenschmidt CE, Peiffer AM, Kraft RA et al (2007) Relating imaging -indices of white matter integrity and volume in healthy older adults. Cereb Cortex 18:433–442
Payne ME, Fetzer DL, MacFall JR et al (2002) Development of a semi-automated method for quantification of MRI gray and white matter lesions in geriatric subjects. Psychiatry Res Neuroimaging 115:63–77
Nebes RD (2002) Longitudinal increase in the volume of white matter hyperintensities in late-onset depression. Int J Geriatr Psychiatry 17:526–530
van Straaten ECW, Fazekas F, Rostrup E et al (2006) Impact of white matter hyperintensities scoring method on correlations with clinical data: the LADIS Study. Stroke 37:836–840
Wen W, Sachdev P (2004) The topography of white matter hyperintensities on brain MRI in healthy 60- to 64-year-old individuals. NeuroImage 22:144–154
Prins ND, van Straaten ECW, van Dijk EJ et al (2004) Measuring progression of cerebral white matter lesions on MRI: visual rating and volumetrics. Neurology 62:1533–1539
Miller DH, Barkhof F, Frank JA et al (2002) Measurement of atrophy in multiple sclerosis: pathological basis, methodological aspects and clinical relevance. Brain 125:1676–1695
Anbeek P, Vincken KL, van Bochove GS et al (2005) Probabilistic segmentation of brain tissue in MR imaging. NeuroImage 27:795–804
Wang Y, Adaly T, Kung SY et al (2008) Quantification and segmentation of brain tissues from MR images: a probabilistic neural network approach. IEEE Trans Image Process 7:1165–1181
Tohka J, Krestyannikov E, Dinov ID et al (2007) Genetic algorithms for finite mixture model based voxel classification in neuroimaging. IEEE Trans Med Imaging 26:696–711
Admiraal-Behloul F, van den Heuvel DMJ, Olofsen H et al (2005) Fully automatic segmentation of white matter hyperintensities in MR images of the elderly. NeuroImage 28:607–617
Sajja BR, Datta S, He R et al (2006) Unified approach for multiple sclerosis lesion segmentation on brain MRI. Ann Biomed Eng 34:142–151
Zhang Y, Brady M, Smith S (2001) Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imag 20:45–57
Deary IJ, Gow AJ, Taylor MD et al (2007) The Lothian Birth Cohort 1936: a study to examine influences on cognitive ageing from age 11 to age 70 and beyond. BMC Geriatr 7:28
MacLullich AM, Ferguson KJ, Deary IJ et al (2002) Intracranial capacity and brain volumes are associated with cognition in healthy elderly men. Neurology 59:169–174
Wardlaw JM, Keir SL, Dennis MS (2003) The impact of delays in computed tomography of the brain on the accuracy of diagnosis and subsequent management in patients with minor stroke. J Neurol Neurosurg Psychiatry 74:77–81
Fazekas F, Barkhof F, Wahlund LO et al (2002) CT and MRI rating of white matter lesions. Cerebrovasc Dis 13(Suppl 2):31–36
Wardlaw J, Ferguson K, Graham C (2004) White matter hyperintensities and rating scales—observer reliability varies with lesion load. J Neurol 251:584–590
Ferguson KJ, MacLullich AMJ, Marshall I et al (2002) Magnetic resonance spectroscopy and cognitive function in healthy elderly men. Brain 125:2743–2749
Mayo Clinic(2008) Analyze 8.1. AnalyzeDirect, Inc. Mayo Clinic. http://www.analyzedirect.com/Analyze/upgrade.asp
Foley JD, van Dam A, Feiner SK et al (1996) Computer graphics: principles and practice in C, 2nd ed. Addison-Wesley Professional
Stockman A, Sharpe LT (2000) Spectral sensitivities of the middle- and long-wavelength sensitive cones derived from measurements in observers of known genotype. Vis Res 40:1711–1737
Wyszecki G, Stiles WS (1982) Color science: concepts and methods, quantitative data and formulae, 2nd edn. John Wiley and Sons, New York
The MathWorks, Inc. (2008) MATLAB reference manual. http://www.mathworks.com/access/helpdesk/help/techdoc/matlab.shtml
Floyd RW, Steinberg L (1976) An adaptive algorithm for spatial grey scale. Proc Soc Inform Displ 17:75–77
Orchard MT, Bouman CA (1991) Color quantization of images. IEEE Trans Signal Process 39:2677–2690
Bland JM, Altman DG (1986) Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307–310
Guillermaud R, Brady M (1997) Estimating the bias field of MR images. IEEE Trans Med Imag 16:238–251
Bedell BJ, Narayana PA, Wolinsky JS (1997) A dual approach for minimizing false lesion classifications on magnetic resonance images. Magn Reson Med 37:94–102
Acknowledgements
This work was funded by “Help the Aged” (now part of AgeUK) and the UK Medical Research Council (funding for MVH) as part of the project “Disconnected Mind: LBC1936”. KJF is funded by an MRC Clinician Scientist Fellowship award to Prof A MacLullich, and JMW in part by SINAPSE (Scottish Imaging Network. A Platform for Scientific Excellence, www.sinapse.ac.uk): a Scottish Funding Council Pooling Initiative. MVH and JMW are members of The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative. Funding from the BBSRC, EPSRC, ESRC and MRC is gratefully acknowledged. The imaging was performed in the SFC Brain Imaging Research Centre (www/sbirc.ac.uk), a SINAPSE Centre. The DICOM to Analyze image format conversion tools used were written by Dr. Paul A. Armitage. The manuscript was proof-read by Miss Natalie A. Royle.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ALEdemo (WMV 10.8 MB)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Valdés Hernández, M.d.C., Ferguson, K.J., Chappell, F.M. et al. New multispectral MRI data fusion technique for white matter lesion segmentation: method and comparison with thresholding in FLAIR images. Eur Radiol 20, 1684–1691 (2010). https://doi.org/10.1007/s00330-010-1718-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-010-1718-6