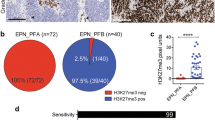
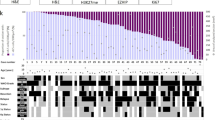
Abstract
Posterior fossa ependymoma comprise three distinct molecular variants, termed PF-EPN-A (PFA), PF-EPN-B (PFB), and PF-EPN-SE (subependymoma). Clinically, they are very disparate and PFB tumors are currently being considered for a trial of radiation avoidance. However, to move forward, unraveling the heterogeneity within PFB would be highly desirable. To discern the molecular heterogeneity within PFB, we performed an integrated analysis consisting of DNA methylation profiling, copy-number profiling, gene expression profiling, and clinical correlation across a cohort of 212 primary posterior fossa PFB tumors. Unsupervised spectral clustering and t-SNE analysis of genome-wide methylation data revealed five distinct subtypes of PFB tumors, termed PFB1-5, with distinct demographics, copy-number alterations, and gene expression profiles. All PFB subtypes were distinct from PFA and posterior fossa subependymomas. Of the five subtypes, PFB4 and PFB5 are more discrete, consisting of younger and older patients, respectively, with a strong female-gender enrichment in PFB5 (age: p = 0.011, gender: p = 0.04). Broad copy-number aberrations were common; however, many events such as chromosome 2 loss, 5 gain, and 17 loss were enriched in specific subtypes and 1q gain was enriched in PFB1. Late relapses were common across all five subtypes, but deaths were uncommon and present in only two subtypes (PFB1 and PFB3). Unlike the case in PFA ependymoma, 1q gain was not a robust marker of poor progression-free survival; however, chromosome 13q loss may represent a novel marker for risk stratification across the spectrum of PFB subtypes. Similar to PFA ependymoma, there exists a significant intertumoral heterogeneity within PFB, with distinct molecular subtypes identified. Even when accounting for this heterogeneity, extent of resection remains the strongest predictor of poor outcome. However, this biological heterogeneity must be accounted for in future preclinical modeling and personalized therapies.




Similar content being viewed by others
References
Araki A, Chocholous M, Gojo J, Dorfer C, Czech T, Heinzl H et al (2016) Chromosome 1q gain and tenascin-C expression are candidate markers to define different risk groups in pediatric posterior fossa ependymoma. Acta Neuropathol Commun 4:88. https://doi.org/10.1186/s40478-016-0349-9
Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D et al (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–474. https://doi.org/10.1038/nature26000
Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B et al (2017) Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31(737–754):e736. https://doi.org/10.1016/j.ccell.2017.05.005
Gojo J, Lotsch D, Spiegl-Kreinecker S, Pajtler KW, Neumayer K, Korbel P et al (2017) Telomerase activation in posterior fossa group A ependymomas is associated with dismal prognosis and chromosome 1q gain. Neuro Oncol 19:1183–1194. https://doi.org/10.1093/neuonc/nox027
Hovestadt V, Remke M, Kool M, Pietsch T, Northcott PA, Fischer R et al (2013) Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol 125:913–916. https://doi.org/10.1007/s00401-013-1126-5
Khatua S, Ramaswamy V, Bouffet E (2017) Current therapy and the evolving molecular landscape of paediatric ependymoma. Eur J Cancer 70:34–41. https://doi.org/10.1016/j.ejca.2016.10.013
Kilday JP, Mitra B, Domerg C, Ward J, Andreiuolo F, Osteso-Ibanez T et al (2012) Copy number gain of 1q25 predicts poor progression-free survival for pediatric intracranial ependymomas and enables patient risk stratification: a prospective European clinical trial cohort analysis on behalf of the Children’s Cancer Leukaemia Group (CCLG), Societe Francaise d’Oncologie Pediatrique (SFOP), and International Society for Pediatric Oncology (SIOP). Clin Cancer Res 18:2001–2011. https://doi.org/10.1158/1078-0432.ccr-11-2489
Korshunov A, Witt H, Hielscher T, Benner A, Remke M, Ryzhova M et al (2010) Molecular staging of intracranial ependymoma in children and adults. J Clin Oncol 28:3182–3190. https://doi.org/10.1200/jco.2009.27.3359
Mack SC, Agnihotri S, Bertrand KC, Wang X, Shih DJ, Witt H et al (2015) Spinal myxopapillary ependymomas demonstrate a warburg phenotype. Clin Cancer Res 21:3750–3758. https://doi.org/10.1158/1078-0432.ccr-14-2650
Mack SC, Pajtler KW, Chavez L, Okonechnikov K, Bertrand KC, Wang X et al (2018) Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature 553:101–105. https://doi.org/10.1038/nature25169
Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stutz AM et al (2014) Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 506:445–450. https://doi.org/10.1038/nature13108
Pajtler KW, Mack SC, Ramaswamy V, Smith CA, Witt H, Smith A et al (2017) The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol 133:5–12. https://doi.org/10.1007/s00401-016-1643-0
Pajtler KW, Wen J, Sill M, Lin T, Orisme W, Tang B et al (2018) Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol. https://doi.org/10.1007/s00401-018-1877-0
Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F et al (2015) Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 27:728–743. https://doi.org/10.1016/j.ccell.2015.04.002
Ramaswamy V, Hielscher T, Mack SC, Lassaletta A, Lin T, Pajtler KW et al (2016) Therapeutic impact of cytoreductive surgery and irradiation of posterior fossa ependymoma in the molecular era: a retrospective multicohort analysis. J Clin Oncol 34:2468–2477. https://doi.org/10.1200/jco.2015.65.7825
Ramaswamy V, Taylor MD (2016) Treatment implications of posterior fossa ependymoma subgroups. Chin J Cancer 35:93. https://doi.org/10.1186/s40880-016-0155-6
Rand V, Prebble E, Ridley L, Howard M, Wei W, Brundler MA, Fee BE, Riggins GJ, Coyle B, Grundy RG (2008) Investigation of chromosome 1q reveals differential expression of members of the S100 family in clinical subgroups of intracranial paediatric ependymoma. Br J Cancer 99:1136–1143. https://doi.org/10.1038/sj.bjc.6604651
Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, Vilo J (2016) g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res 44:W83–W89. https://doi.org/10.1093/nar/gkw199
Ruda R, Reifenberger G, Frappaz D, Pfister SM, Laprie A, Santarius T et al (2018) EANO guidelines for the diagnosis and treatment of ependymal tumors. Neuro Oncol 20:445–456. https://doi.org/10.1093/neuonc/nox166
Ryall S, Guzman M, Elbabaa SK, Luu B, Mack SC, Zapotocky M, Taylor MD, Hawkins C, Ramaswamy V (2017) H3 K27M mutations are extremely rare in posterior fossa group A ependymoma. Childs Nerv Syst 33:1047–1051. https://doi.org/10.1007/s00381-017-3481-3
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N et al (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378
Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C et al (2012) Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22:425–437. https://doi.org/10.1016/j.ccr.2012.08.024
Taylor MD, Poppleton H, Fuller C, Su X, Liu Y, Jensen P et al (2005) Radial glia cells are candidate stem cells of ependymoma. Cancer Cell 8:323–335. https://doi.org/10.1016/j.ccr.2005.09.001
Thompson YY, Ramaswamy V, Diamandis P, Daniels C, Taylor MD (2015) Posterior fossa ependymoma: current insights. Childs Nerv Syst 31:1699–1706. https://doi.org/10.1007/s00381-015-2823-2
Ward S, Harding B, Wilkins P, Harkness W, Hayward R, Darling JL, Thomas DG, Warr T (2001) Gain of 1q and loss of 22 are the most common changes detected by comparative genomic hybridisation in paediatric ependymoma. Genes Chromosomes Cancer 32:59–66. https://doi.org/10.1002/gcc.1167
Witt H, Mack SC, Ryzhova M, Bender S, Sill M, Isserlin R et al (2011) Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell 20:143–157. https://doi.org/10.1016/j.ccr.2011.07.007
Acknowledgements
VR is supported by operating funds from the Canadian Institutes of Health Research, American Brain Tumor Association, the Garron Family Cancer Center, Meagan’s Walk, b.r.a.i.n.child, the Brain Tumor Foundation of Canada and a Collaborative Ependymoma Research Network (CERN) basic science fellowship. FMGC is supported by the Stephen Buttrum Brain Tumor Research Fellowship, granted by Brain Tumor Foundation of Canada. MZ is supported by Garron Family Cancer Center Fellowship, Meagan’s Walk Neuro-Oncology Fellowship and Restracomp from Research Training Center at The Hospital for Sick Children. MDT is supported by funds from the Garron Family Chair in Childhood Cancer Research at The Hospital for Sick Children and The University of Toronto, and operating funds from the National Institutes of Health (R01CA159859 and R01CA148699), and the Pediatric Brain Tumor Foundation. EB is supported by funds from the Garron Family Chair in Childhood Cancer Research at The Hospital for Sick Children and The University of Toronto. This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748 to Memorial Sloan Kettering Cancer Center. KWP is supported by a CERN Research Fellowship.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Florence M. G. Cavalli, Jens-Martin Hübner, and Tanvi Sharma are co-first authors.
Marcel Kool and Vijay Ramaswamy are co-senior authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
401_2018_1888_MOESM1_ESM.eps
Supplemental Fig. 1A–F: Spectral clustering of PFB at k = 2–7. Heatmap of the similarity matrix obtained by methylation data on the 212 primary PFB samples. Yellow represents more similar and red represents dissimilar. Color bar at the top represents the final PFB1–5 cluster assignation of the samples (EPS 38612 kb)
401_2018_1888_MOESM2_ESM.eps
Supplemental Fig. 2: Genomic comparison of PFB subtypes. A) Comparison of subtype assignments between spectral clustering and t-SNE analysis. Relationships (indicated by the grey bars between columns) between the groups of samples obtained by t-SNE analysis (3 to 5 clusters) and the groups obtained by spectral clustering (SpC, 2–5 clusters). B) Density plot of overall promoter methylation levels across the five PFB subtypes (EPS 2173 kb)
401_2018_1888_MOESM3_ESM.eps
Supplemental Fig. 3: List of differentially regulated pathways across PFB subtypes. Enrichment of specific pathways within the PFB subtypes using significantly differentially expressed genes (adjusted p < 0.05). No pathway were identified for PFB2 and 3, since those two subtypes had a low number of significantly expressed genes (EPS 1839 kb)
401_2018_1888_MOESM4_ESM.eps
Supplemental Fig. 4: Unsupervised clustering of broad copy-number events across PFB subtypes. A) t-SNE analysis of broad copy-number profiles across 212 PFB ependymomas with corresponding methylation t-SNE subtype assigned. B) Unsupervised consensus clustering of broad copy-number events using negative matrix factorization (NMF) from k = 2 to k = 6 and a plot of the cophenetic coefficient (EPS 31557 kb)
401_2018_1888_MOESM5_ESM.eps
Supplemental Fig. 5: Arm-level cytogenetic events at k = 4 PFB subtypes from t-SNE analysis. A) Frequency and significance of arm-level gains and losses across the 4 PFB subtypes. Darker bars show significant arm-level events (q value ≤ 0.1, Chi-squared test). B) Frequency and significance of whole chromosome gain and losses across the four PFB subtypes. Darker bars show significant arm-level events (q value ≤ 0.1, Chi-squared test) (EPS 1472 kb)
401_2018_1888_MOESM6_ESM.eps
Supplemental Fig. 6: Clinical characteristics of PFB subtypes. A) Progression-free and B) overall survival across PFB subtypes. P values determined using the log-rank test. C) Pie chart of the administration of upfront radiation across PFB subtypes and D) Pie chart of the incidence of gross total resection across PFB subtypes. P values determined using the Fisher’s exact test. Overall survival stratified by E) 1q gain and F) 13q status. p values determined using the log-rank test (EPS 2421 kb)
Rights and permissions
About this article
Cite this article
Cavalli, F.M.G., Hübner, JM., Sharma, T. et al. Heterogeneity within the PF-EPN-B ependymoma subgroup. Acta Neuropathol 136, 227–237 (2018). https://doi.org/10.1007/s00401-018-1888-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-018-1888-x