Abstract
BACKGROUND AND PURPOSE: With appropriate selection, temporal lobe epilepsy is potentially curable with surgical intervention achieving seizure freedom in ∼80% of individuals. MR imaging−based identification of MTS remains central to the selection process but currently relies on qualitative visual analysis. We sought to determine if new ultrastructural hippocampal details seen on 3T MR imaging had histopathologic correlates and whether these could serve as a useful tool in MTS identification.
MATERIALS AND METHODS: Patients undergoing selective anterior temporal lobectomy (n = 5) were scanned using 3T MR imaging preoperatively. En bloc resections were rescanned and subsequently prepared for histopathologic analysis of all hippocampal layers in the CA1–3 regions. Using a newly identified landmark from 3T FSTIR coronal images in 20 patients with histologically confirmed MTS, blinded studies compared ipsilateral and contralateral sides to generate threshold measurements for application in a fast quantitative analysis tool.
RESULTS: Histopathologic analysis and correlation with 3T imaging of en bloc resections identified the low-intensity signal as the stratum lacunosum. MTS was associated with extensive gliosis throughout the CA1–3 regions, with loss of tissue thickness in the stratum pyramidale most pronounced in CA1. Fast quantitative analysis by using the stratum lacunosum as a landmark provided a test that identifies MTS with a SN of 70% and SP of 85%.
CONCLUSIONS: Here we delineated ultrastructural hippocampal details seen on 3T MR imaging in both the in vivo and ex vivo setting, correlating these with histopathologic features consistent with MTS, and provided preliminary data suggesting their utility in the development of a quantitative analysis assessment tool for application in surgical-candidate selection.
Abbreviations
- CA
- cornu ammonis
- EEG
- electroencephalography
- FLAIR
- fluid-attenuated inversion recovery
- FSTIR
- fast short tau inversion recovery
- GFAP
- glial fibrillary acidic protein
- L
- left
- LFB-HE
- Luxol fast blue/hematoxylin-eosin
- mTLE
- mesial temporal lobe epilepsy
- MTS
- mesial temporal sclerosis
- R
- right
- SN
- sensitivity
- S
- stratum
- SNR
- signal-to-noise ratio
- SP
- specificity
- STIR
- short tau inversion recovery
MTLE is most frequently associated with hippocampal sclerosis, and, though often refractory to medical therapy, has been shown to be amenable to surgery, with up to 80% of individuals being able to attain seizure freedom after resection of the mesial temporal structures and variable amounts of temporal neocortex.1 Identification of surgical candidates is based on EEG recordings, neuropsychological testing, and MR imaging analysis. The main clinical goal of imaging in mTLE is the identification of MTS or an alternative structural lesion located in the mesial temporal lobe because identification of either is associated with a positive surgical outcome.2,3 Images acquired with 3T MR imaging provide significantly improved detail of hippocampal anatomy, which is less likely to be visualized by 1.5T systems.4 Visualization of these structural details may provide stronger evidence of hippocampal atrophy, increasing the diagnostic certainty of MTS and the ability to correlate other preoperative investigations to the presence of an epileptogenic lesion, thereby reducing diagnostic uncertainty and thus the possible need for further investigations.
Identification of MTS in patients with suspected temporal lobe epilepsy has routinely relied on qualitative assessment of hippocampal structures through expert visual analysis.5 The presence of MTS is dependent on the presence of 2 abnormalities involving the hippocampus: 1) atrophy, and 2) increased T2 signal intensity. STIR sequences have been effective in revealing atrophic changes, while T2-weighted FLAIR sequences have been effective in displaying T2 signal-intensity increases associated with MTS. In an attempt to improve the quantitative analysis of MTS, volumetry, T2 relaxometry, and spectroscopy have been applied with some success.6 These methods require postprocessing analysis that has limited universal acceptance. Improved image quality can be achieved when moving from 1.5 to 3T MR imaging systems as a result of the linear increase in SNR associated with higher field strengths.7 The higher SNR afforded by imaging at 3T can be used to increase spatial resolution while maintaining the same SNR as images acquired at a lower spatial resolution at 1.5T. This increase in spatial resolution has improved image quality and led to new identification of poorly seen structural details (Fig 1).4,8
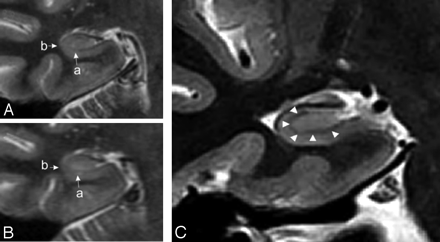
Right hippocampus in a coronal section showing anatomic detail in 3T MR imaging (A) versus 1.5T MR imaging (B). Small arrows indicate the hypoattenuated region later determined to represent the stratum lacunosum (a) and lateral margin (b), both of which are significantly more distinguishable on 3T MR imaging. C, Right hippocampus shows ultrastructural detail determined on 3T MR imaging (T2 FSTIR). Coronal view of the right hippocampus shows distances that can be measured by using improved neuroanatomic detail. Arrowheads point to a consistently observed band seen on coronal sections in 3T MR imaging.
Histopathologic correlation with newly identified structures on 3T MR imaging is important for assessing the significance of emergent data in the context of a pathophysiologic process. MTS is a process that reflects neuronal loss and gliosis thought to be in part genetic9 and in part secondary to repeated insults from hyperexcitability and recurrent seizure activity originating in the temporal lobe.10 In our center, we had the unique opportunity to study hippocampi with MTS following neurosurgical en bloc resection in a highly selective patient population. Here we examined ex vivo hippocampal sections by using 3T MR imaging and histopathologic analysis and compared preoperative imaging findings to determine if there is a correlation with atrophy and/or gliosis of specific hippocampal regions. Because 3T MR imaging provides improved imaging of the internal architecture of the hippocampus, we further sought to determine if measuring loss of height within the CA1, 2, and 3 regions on an image corresponding to the head of the hippocampus could be correlated with observed histologic changes and could be applied as a fast quantitative assessment tool for MTS. Through this investigation, we show loss-of-height measurements taken from a single cross-sectional image not only correlate with the degree of MTS (primarily loss of CA1 neurons), a feature obtained solely from 3T imaging, but they also have potential application as a simple yet robust preoperative assessment of MTS in patients with suspected mTLE.
Materials and Methods
Subjects
Patients with temporal lobe epilepsy underwent routine preoperative assessments as per institution protocols (University Health Network, Toronto Western Hospital, Toronto, Canada). Briefly, patients were admitted for EEG–video telemetry, during which time they underwent neuropsychological evaluation. Language dominance was determined by functional MR imaging, and the Wada test was performed only for those in whom it was suspected that the contralateral hemisphere would not support memory.11,12 None of the patients in this series underwent intracranial implantation of subdural strip and depth electrodes. Preoperative assessment was based on qualitative MR imaging analysis by using a 3T magnet (Signa HDx; GE Healthcare, Milwaukee, Wisconsin) to identify hippocampal sclerosis by using a 3T epilepsy protocol (sagittal T1 FLAIR, coronal T2 FLAIR, coronal 512 FSTIR: TR, 6500 ms; TE, 40.9 ms; echo-train length, 12; display FOV, 22 × 22 cm; matrix, 512 × 512).
Interpretation of clinical studies and selection of potential surgical candidates were performed following weekly multidisciplinary case conferences. Informed consent was obtained for anterior temporal lobectomy of the affected side as well as participation in human tissue research. 3T MR imaging was repeated for neuronavigation on the morning of surgery. For this study, 20 patients were identified from cases of histologically confirmed MTS completed during a 2-year period, with 5 separate consecutive patients selected during a 5-month period for ex vivo analysis (3 of whom were also included in the previously mentioned fast quantitative analysis study).
Identifying Hippocampal Atrophy by Using Fast Quantitative Analysis
On the basis of work completed by Bhuta et al,8 measurements generated by receiver operating characteristic analysis by using 3T MR imaging anatomic detail have been proposed for a fast quantitative method of assessing hippocampal sclerosis. Briefly, through comparison of cumulative measurements taken at the superior, inferior, and lateral hippocampal margins (Fig 2) in 25 cases of unilateral MTS versus healthy controls, hippocampal atrophy could be correctly identified with an SN and SP of 64% and 96%, respectively. In our study, measurements from the superior, inferior, and lateral margins were acquired for 20 cases of confirmed MTS on the basis of 3T MR imaging (coronal 512 FSTIR and sagittal T1 for localizing the head of the hippocampus) by using the Advantage Workstation (AW4.2_07 sdc; GE Healthcare). Analysis was performed bilaterally by an individual blinded to the side of MTS. Means of cumulative values were compared between MTS and the contralateral side by using the Student t test. SN and SP values for determining hippocampal atrophy were calculated on the basis of measurements falling within 1 SD of the normal (contralateral) side.
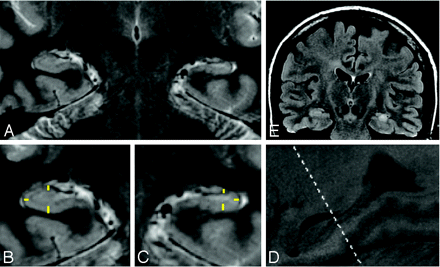
Sample images from patient 4 depicting a coronal section showing left-sided hippocampal sclerosis (A) with enlarged images of the right (B) and left (C) sides. Note the yellow markers used to depict the superior, inferior, and lateral measurements (sample only, actual images with measurements from Advantage Workstation could not be exported). D, The enlarged image depicts the hippocampus in a sagittal section with the head of the hippocampus indicated by the dashed line, used to determine the appropriate coronal image from which measurements could be taken. E, Corresponding coronal T2 FLAIR image shows increased signal intensity on the left side, a radiographic marker for a sclerotic process suggestive of gliosis.
Ex Vivo Scanning by Using 3T MR Imaging
Surgical specimens resected en bloc were immediately fixed in 10% neutral-buffered formalin and scanned within 2–3 weeks by using 3T MR imaging (Signa HDX) and a customized solenoidal radio-frequency coil (diameter = 3 cm). Specimens were suspended in 50-mL polypropylene tubes (BD Falcon; BD Biosciences, San Jose, California) with the anteroposterior axis in the upright position. Vibrational motion artifacts were minimized by elimination of air pockets and maintaining the upright position with rubber stoppers at both ends. Coronal images were obtained with the following parameters: T2 fast relaxation fast spin-echo; TR, 2500 ms; TE, 102 ms; echo-train length, 27; section thickness, 1.0 mm with a 0.5-mm gap; FOV, 4 × 4 cm; matrix, 256 × 256, NEX, 36; phase FOV, 0.8 × 0.8 cm. Image acquisition was focused in regions where the hypointense band, later determined to represent the stratum lacunosum (Fig 1), was most apparent on a 3-plane localizer. Images were sent to a server for storage and retrieval.
Histopathology: Light Microscopy and Image Analysis
For histopathologic analysis, the entire hippocampus was sectioned at 2-mm intervals and entirely submitted for histologic analysis. Specimens were sectioned from anterior to posterior in a coronal plane that was identical to the plane of ex vivo imaging. Samples were paraffin-processed and embedded. For light microscopy, 5-μm-thick sections were stained with an LFB-HE combination. Immunohistochemistry for GFAP (prediluted antibody; Ventana Medical Systems, Oro Valley, Arizona) was performed on 5-μm-thick sections by using an automated stainer (Ventana Medical Systems). All sections were counterstained with hematoxylin. Negative controls were derived from brain autopsies with unremarkable findings. Neurosurgical cases showing extensive astrogliosis derived from cerebral trauma served as positive controls.
Image Analysis
Virtual Microscopy.
All histologic slides were scanned (original magnification ×200) on a ScanScope CS digital whole-slide scanner for subsequent analysis by using the ImageScope, Version 9.1 software (Aperio Technologies, Vista, California). The Positive Pixel Count Tool (Aperio Technologies) was used to analyze GFAP and LFB-HE slides. For the GFAP immunostain, brown color was detected by using a hue value = 0.66, while for the LFB-HE stain, a hue value = 0.1 was used to highlight blue color. The presence of gliosis was evaluated by immunohistochemistry for GFAP on standardized sections from the anterior, central, and posterior hippocampus. The luminosity was assessed as previously described13 and compared with age-matched control cases. To quantify the findings, GFAP immunoreactivity was measured in Photoshop Elements 5.0 (Adobe Systems, San Jose, California) by using the histogram tool. The average luminosity of images was measured, and the staining intensity was expressed as [1000] / [luminosity value]. An average normal intensity was derived from the reference cases, and the study cases were compared with this average, expressed as a percentage. LFB-HE stains were performed to assess the extent of loss of myelinated fibers. Scoring was performed analogous to the GFAP immunostains.
The boundaries of the anatomic subfields of the hippocampal formation were assessed on LFB-HE stains. Region CA3 was easily distinguished from the adjacent region CA4 by the location of the neighboring dentate gyrus. The distinction between CA1 and CA2 is based on the perikaryal size of the pyramidal cells, a difference that was still notable even in the specimens with extensive sclerosis.
Pixel Count.
Fifteen boxes were placed within each subsection of CA1, 2, and 3, in the stratum radiatum (original magnification ×8) in the location of the band of interest. Each box measured 250 × 250 μm. The Positive Pixel Count Tool was then applied, generating average positivity values for each region. The positivity value is expressed as a ratio of positive pixels detected to total area analyzed within each enclosed area.
Thickness Measurements.
The ruler tool in ImageScope was used to measure the thickness of each layer within the hippocampus. Within each subsection of the hippocampus, CA1, −2, and −3, the following 4 areas were measured: stratum pyramidale, stratum radiatum, stratum lacunosum, and a measurement from the stratum oriens (outer edge of the hippocampus) to the superficial layer of the stratum lacunosum. For each area, 5 measurements were taken and averaged.
Results
Patient Demographics
Of the 20 patients with histologically confirmed MTS who underwent hippocampal measurements, 12 were left-sided. All were adults (>18 years of age), 13 female, with seizures refractory to medical management. All had undergone anterior temporal lobectomy at the Toronto Western Hospital from October 2005 to February 2009. All patients had evidence of hippocampal sclerosis identified by preoperative expert neuroradiologic review with MTS diagnosis determined following histopathologic assessment of resected surgical specimens by institution-based neuropathologists. Patients that were MR imaging “healthy” or in whom diffuse MTS was reported were not included, in an attempt to optimize measurement validity.
For the ex vivo study, data for the 5 patients are summarized in the Table. Briefly, all were adults (age range, 24–51 years) and female (random occurrence) and had undergone anterior temporal lobe resection between October 2006 and February 2007. On the basis of preoperative neuroradiologic qualitative assessment, 4 of 5 patients had features of MTS, 3 of which were right-sided. One case was classified as diffuse temporal dysplasia (patient 1). EEG localized seizure activity to the anterior temporal lobe ipsilateral to the involved side, with 1 case demonstrating bilateral activity despite a predominance of seizures being initiated in the anticipated region (patient 5). Radiologic and EEG data were concordant with neuropsychological testing. Seizure history ranged from 5 to 44 years and had been poorly controlled by medical management before the operation. All were seizure-free at 1 year postoperatively.
Patient demographics (ex-vivo and histopathologic analysis)
Identification of Hippocampal Atrophy by Using 3T MR Imaging Anatomic Detail for Fast Quantitative Analysis
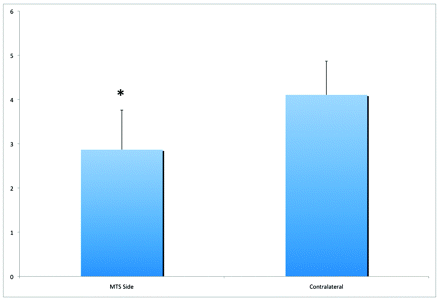
Measurements were based on the cumulative value of superior, inferior, and lateral hippocampal margins for each side, on the basis of a region defined by the low-attenuation band seen in the coronal plane on 3T MR imaging. In comparison with the contralateral controls, MTS hippocampi were significantly smaller, with a mean cumulative value of 2.9 ± 0.9 mm versus 4.1 ± 0.8 mm (P < .001, Fig 3). Consistent with this, values were significantly different between the MTS and contralateral sides for each separate measurement (1.0 ± 0.48 versus 1.6 ± 0.53 lateral**; 0.7 ± 0.25 versus 0.9 ± 0.26 superior*; 1.2 ± 0.37 versus 1.6 ± 0.32 inferior**; *P < .01, **P < .001). The utility of a fast quantitative analysis is based in part on a test with high SN and SP. Using a threshold value of 3.3 mm, this measurement tool has an SN and SP of 70% and 85%, respectively. This threshold was based on a cumulative measurement value falling within 1 SD of the contralateral control (ie, values ≤3.3 mm indicated MTS).
Cumulative hippocampal measurements of superior, inferior, and lateral margins comparing the MTS side with the contralateral internal control. Values are presented in millimeters (mean ± SD, P < .001; n = 20).
Ultrastructural Detail of the Hippocampus from 3T MR Imaging Reveals the Stratum Lacunosum
To identify the low-intensity signal band visible within the hippocampus on 3T MR imaging, we imaged ex vivo hippocampal specimens with the same 3T MR imaging system and histopathologic analysis performed on the corresponding coronal sections. Figure 4 shows corresponding sections from 1 case (patient 4), permitting identification of the stratum lacunosum via overlay analysis.
A, Postoperative ex vivo MR image of the hippocampal specimen in a coronal section. B, Scanned image of corresponding histologic slide (× 20 magnification) with CA1, CA2, and CA3 regions marked accordingly. Overlay of both images clarifies structural MR imaging information, such as the dark band that was ultimately identified as the stratum lacunosum (arrow).
Ultrastructural Hippocampal Anatomy Observed on 3T MR Imaging Is Associated with Decreased Tissue Thickness in Specific Hippocampal Layers and Increased Gliosis Throughout the CA1, −2, and −3 Regions
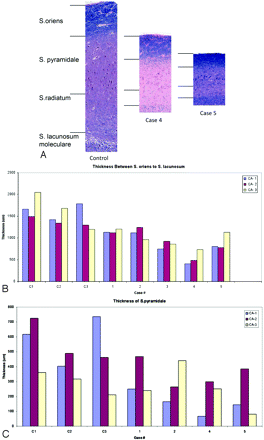
A small number of en bloc surgical resections were scanned postoperatively and subsequently processed for histopathologic analysis, to correlate a pathophysiologic process with the decreased hippocampal measurements determined in patients with confirmed MTS. For all 5 cases examined, MTS was associated with neuronal loss, particularly in the CA1 region, as demonstrated by decreased thickness across the hippocampal layers (Fig 5A). In the 2 most dramatic examples, neuronal loss and decreased tissue thickness were greatest between the outer margin (stratum oriens) and stratum lacunosum (low-intensity signal band observed on 3T MR imaging), localized to the stratum pyramidale in CA1 in particular (Fig 5B, -C).
A, Comparison of layer thickness within the CA1 between the control and 2 cases of MTS. Note the marked reduction in thickness, particularly in the stratum pyramidale and radiatum in cases 4 and 5. B, Thickness measurement of the distance between the stratum oriens and stratum lacunosum in all 3 CA regions. Again note the marked size reduction in cases 4 and 5 compared with all 3 controls (C1, C2, C3). C, Specific thickness measurements of the stratum pyramidale in regions CA1, -2, and -3 with marked size reduction apparent in the CA1 region of cases 4 and 5.
Gliosis is another hallmark of mesial temporal sclerosis. As expected, there was strong immunoreactivity for GFAP staining in the CA1, −2, and −3 regions of hippocampal specimens from the en bloc surgical resections (Fig 6), a finding consistent with MTS. Together, the combination of neuronal loss, gliosis, and decreased tissue thickness suggests a structural process underlying the reduced hippocampal measurements derived from neuroanatomic details unique to 3T MR imaging.
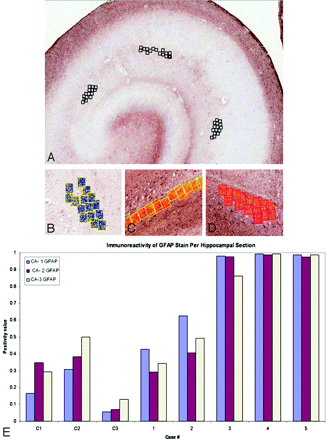
A, Methodology for Positive Pixel Count Tool. Placement of pixel count boxes in 3 sections of the hippocampus: CA1, -2, and -3. B−D, Positive Pixel Count Tool: immunoreactivity is converted to blue (LFB-HE) or red and orange (GFAP stain). E, Analysis of immunoreactivity of the GFAP stain within each CA region of the hippocampal specimens. Note marked increase in GFAP staining in all MTS cases compared with control cases.
Discussion
Epilepsy is prevalent in 0.6% of the population, and poorly controlled by medications in almost 30% of these patients.14 In appropriate surgical candidates, there is an approximately 80% chance of being seizure-free at 1 year postoperatively, with outcomes being influenced by several factors including preoperative hippocampal sclerosis and seizure localized to the anterior temporal lobe.15 Patient selection, therefore, requires an interdisciplinary approach with multiple assessment modalities. Visual (ie, qualitative) analysis identifying hippocampal atrophy (surrogate for the pathologic diagnosis of MTS) on radiographic imaging remains a cornerstone of this process. While methodologies exist to perform quantitative analysis (eg, volumetry and T2 relaxometry, reviewed by Knowlton16), there is no universal acceptance of their use due to concerns over lengthy operator time and significantly variable software programs. It is promising that with the advent of new imaging modalities, such as 3T MR imaging, previously unrecognized ultrastructural details may provide new means of assessing patients with temporal lobe epilepsy for radiologic evidence of MTS. In this study, we applied a previously reported fast quantitative analysis method derived from an anatomic detail unique to 3T MR imaging as a means to identify MTS, highlighting the potential utility of this tool. Furthermore, we identified this new landmark as the stratum lacunosum and provided a pathophysiologic correlate for the structural differences detected by our measurement tool through ex vivo imaging and histopathologic analysis showing neuronal loss and gliosis.
Our application of a fast quantitative analysis tool to confirmed cases of MTS supports the work of Bhuta et al,8 in which select measurements at the head of the hippocampus by using landmarks seen on 3T MR imaging were used to identify atrophy. Here we determined that patients with MTS have significantly diminished hippocampal tissue height at the superior, inferior, and lateral margins, regions bordered by a band of low-intensity signal on coronal FSTIR sequences. This observation was even more robust when comparison of the cumulative values was made. We subsequently defined a threshold measurement of ≤3.3 mm (1 SD below the cumulative mean), which identified MTS with an SN and SP of 70% and 85%, respectively. While Bhuta et al similarly identified 3.4 mm as a threshold measurement, the SN and SP of their test differed (SN 64% and SP 96%). Selection of control measurements may account for the variation between these 2 studies because we had an internal control using the contralateral hippocampus, while the earlier study compared MTS measurements with age-matched healthy controls from non-temporal lobe disease studies. It is difficult to determine if 1 type of control has greater value because the contralateral measurement can help lateralize a disease process that may be bilateral,17 while age-matched healthy individuals provide a measure against which absolute disease thresholds can be set. Future studies could address this issue by directly comparing measurements of age-matched healthy individuals with those acquired in this study.
Implementing a universally accepted quantitative analysis tool has been difficult to date. The tool described in this study has limitations as well. The use of 1 SD in generating a threshold measurement represents a value that optimized test utility because extending the range of normal measurements (eg, 2 SDs) significantly increased the false-negatives, thereby affecting SN. Given that these measurements are quantifying a sclerotic process in terms of millimeters of lost tissue height and that bilateral disease may be present, the use of a more stringent threshold value seems fitting. Moreover, our tool will not replace visual analysis as a single mode of assessment without further improvement, given that visual analysis has a test SN and SP of 86% and 83%, respectively.18 It may, however, enable more novice radiologists or indeterminate imaging to benefit from additional but time-efficient evaluation.
To characterize the low-intensity signal-landmark and correlate altered hippocampal measurements with a pathophysiologic process, we scanned and analyzed a small subset of surgical en bloc resections obtained during a 5-month period, for regions of neuronal loss and gliosis. With the exception of all 5 patients being female (random occurrence), patient demographics were comparable and all patients were seizure-free at 1 year postoperatively. Similar to the in situ preoperative imaging, a white matter tract was consistently identified on ex vivo scans as a low-intensity signal band, which we subsequently identified as the stratum lacunosum through overlay analysis (Figs 1 and 4). Determining the histopathologic correlates of neuroimaging has become increasingly necessary as newly visualized radiologic features are used for diagnostics. It has been shown that T2 relaxation time is negatively correlated with neuronal loss (decreased NeuN staining) in gray matter but not in white matter tracts by using 1.5T MR imaging.19
Using 3T MR imaging, Eriksson et al4 analyzed T2-weighted images from patients with hippocampal sclerosis with a sequence designed to reduce head motion artifact (periodically rotated overlapping parallel lines with enhanced reconstruction) and correlated a band of myelinated tissue (seen as the band of low-intensity signal) with the stratum lacunosum and radiatum, supporting the findings in our study. In the 4 patients studied, they noted an abrupt transition to atrophy, neuronal loss, and gliosis in the CA1 region in particular. In contrast, we found that gliosis is extensive throughout the CA1, -2, and -3 regions, with loss of tissue thickness in the stratum pyramidale most pronounced in CA1. In our study, we had the advantage of being able to directly compare ex vivo scans with histopathologic specimens, eliminating discrepancies caused by tissue distortion when comparing in vivo preoperative scans with ex vivo specimens as in other studies. Indeed, the decreased tissue thickness on histopathologic analysis (MTS values, 500–1200 μm versus 1400–1700 μm for controls) is consistent with decreased tissue thickness calculated by using the stratum lacunosum landmark on 3T studies (200–1300 μm—superior margin measurement range), supporting a process that correlates with radiographic features of MTS. While ex vivo scans with 9.4T research magnets have detailed exceptional hippocampal structure,20 these are not clinically available systems. Here we have shown for the first time, ultrastructural details of hippocampi in both the in vivo and ex vivo setting by using 3T MR imaging, a magnet strength now available at many centers, and we have correlated this with histopathologic features consistent with MTS.
In light of data suggesting 5 distinct MTS patterns with the classic pattern described as severe cell loss in CA1 and moderate loss in all other subfields,21 it is possible that our measurement technique will lead to bias toward identifying specific subsets of patients with MTS. Indeed, the most striking difference between ipsilateral and contralateral tissue measurements occurred in patients 4 and 5, both of whom also demonstrated the greatest neuronal loss in the CA1 region. A particularly key group of patients to identify, however, are those considered MR imaging “healthy.” Although we did not apply our measurement tool to a complete group of these patients, it is interesting that in 1 of 2 “MR imaging-healthy” patients who were excluded from the fast quantitative analysis study, post hoc evaluation showed the measurement tool would have accurately predicted the ultimate histologic diagnosis of MTS. Nevertheless, it seems unlikely that a test SN of 70% would be adequate for screening MR imaging–healthy patients at this time. Most interesting, these patients have been described as having a syndrome of paradoxic temporal lobe epilepsy with neuronal loss less pronounced in CA1 compared with a region of maximal loss in CA4.22 It may be that new ultrastructural features specific to this group or unique imaging modalities will be required.
Conclusions
While these studies together suggest temporal lobe epilepsy represents a spectrum of histopathologic features, it is promising that diminished hippocampal measurements generated by using 3T MR imaging ultrastructural details are consistent with characteristic MTS features. Moreover, the possibility of applying new radiographic landmarks, such as the stratum lacunosum identified in this study, for use with a fast quantitative analysis tool, may help to more accurately identify patients with uncontrolled seizures who would benefit from surgical intervention. Because hippocampal sclerosis on MR imaging remains associated with excellent seizure outcome after temporal lobectomy,15 this remains a field requiring improved tests for optimizing diagnostic accuracy.
Acknowledgments
We thank Brian Rutt, MD, (Stanford University) for generously providing the coil for ex vivo specimen MR imaging.
References
- Received January 16, 2010.
- Accepted after revision March 16, 2010.
- Copyright © American Society of Neuroradiology