Abstract
SUMMARY: MRS, including single-voxel spectroscopy and MR spectroscopic imaging, captures metabolites in high-grade gliomas. Emerging evidence indicates that 7T MRS may be more sensitive to aberrant metabolic activity than lower-field strength MRS. However, the literature on the use of 7T MRS to visualize high-grade gliomas has not been summarized. We aimed to identify metabolic information provided by 7T MRS, optimal spectroscopic sequences, and areas for improvement in and new applications for 7T MRS. Literature was found on PubMed using “high-grade glioma,” “malignant glioma,” “glioblastoma,” “anaplastic astrocytoma,” “7T,” “MR spectroscopy,” and “MR spectroscopic imaging.” 7T MRS offers higher SNR, modestly improved spatial resolution, and better resolution of overlapping resonances. 7T MRS also yields reduced Cramér-Rao lower bound values. These features help to quantify D-2-hydroxyglutarate in isocitrate dehydrogenase 1 and 2 gliomas and to isolate variable glutamate, increased glutamine, and increased glycine with higher sensitivity and specificity. 7T MRS may better characterize tumor infiltration and treatment effect in high-grade gliomas, though further study is necessary. 7T MRS will benefit from increased sample size; reductions in field inhomogeneity, specific absorption rate, and acquisition time; and advanced editing techniques. These findings suggest that 7T MRS may advance understanding of high-grade glioma metabolism, with reduced Cramér-Rao lower bound values and better measurement of smaller metabolite signals. Nevertheless, 7T is not widely used clinically, and technical improvements are necessary. 7T MRS isolates metabolites that may be valuable therapeutic targets in high-grade gliomas, potentially resulting in wider ranging neuro-oncologic applications.
ABBREVIATIONS:
- CRLB
- Cramér-Rao lower bound
- 2D L-COSY
- 2D localized correlated spectroscopy
- FID
- free induction decay
- GABA
- gamma-aminobutyric acid
- GBM
- glioblastoma
- Gln
- glutamine
- Glu
- glutamate
- Gly
- glycine
- GPC
- glycerophosphocholine
- GSH
- glutathione
- 2HG
- D-2-hydroxyglutarate
- HGG
- high-grade glioma
- Lac
- lactate
- LGG
- low-grade glioma
- MRSI
- MR spectroscopic imaging
- PC
- phosphocholine
- PRESS
- point-resolved spectroscopy sequence
- RF
- radiofrequency
- SVS
- single-voxel spectroscopy
- SAR
- specific absorption rate
- SASSI
- Semi-Adiabatic Spectral-spatial Spectroscopic Imaging
- tCho
- total choline
- WHO
- World Health Organization
High-grade gliomas (HGGs) are the most lethal and common types of adult brain cancer, including both World Health Organization (WHO) grade III astrocytomas and grade IV glioblastomas (GBMs).1 With >10,000 new cases every year in the United States, GBM accounts for 60% of primary adult brain tumors.2 Even with surgical resection, chemotherapy, and radiation, GBM is associated with a median survival time of 15 months.3 In light of the deadliness and prevalence of HGGs, considerable research has been devoted to uncovering their metabolic profiles, detected by noninvasive MRS techniques such as single-voxel spectroscopy (SVS) and multiple-voxel MR spectroscopic imaging (MRSI).4,5 Unless otherwise stated, MRS in this document always refers to 1H-MRS. In this article, we will primarily concentrate on MRS and MRSI but will also occasionally refer to additional imaging methods that can be performed with clinical MR imaging scanners or widely available MR imaging technology and can complement spectroscopic imaging.
MRS characterizes each metabolite on the basis of its unique set of chemical shifts and has been shown to capture up to 17 metabolites, including but not limited to Cho, Cr, glutamate (Glu), glutamine (Gln), lactate (Lac), lipids, mIns, and NAA (Fig 1).6 It can also detect additional metabolites more associated with gliomas such as glycine (Gly) and D-2-hydroxyglutarate (2HG), an oncometabolite produced in mutant isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) tumors.7⇓-9 Most studies on these metabolites have involved MRS at 1.5T and 3T.10⇓-12 Although 3T MRS has value, it is limited in several ways.13 One shortcoming is its limited ability to quantify metabolites with overlapping peaks.14 Another is its low spatial resolution, which detracts from tumor characterization.15 These drawbacks have prompted more focus on higher-field-strength MR imaging systems.16
A, An MR spectrum generated at 7T from the occipital lobe (predominantly gray matter) of a healthy volunteer is shown (TR/TE = 2000/30.5 ms; number of averages = 32). The real part appears in blue and includes metabolite labels. The same image processed with LCModel is shown below and contains quantified metabolite values. Instead of capturing the increased resolution attainable at higher field strengths, these data are intended to primarily show a very clear spectrum from a normal brain. This was a 3 × 3 × 3 SVS acquisition in 1 minute obtained using an SASSI sequence. In contrast, note a single-voxel 1H-MRS spectrum (TR/TE = 3000/23 ms; number of averages = 8) from a patient with a grade III astrocytoma located in the left parietal region and having an IDH-mutant genotype showing various metabolites (B). The patient was scanned on a 7T whole‐body MR imaging scanner equipped with a single transmit/32‐channel receiver array head coil. Tissue infiltrated with gliomas such as the astrocytoma in B results in spectra with different metabolic characteristics than the normal tissue in A. Lip indicates lipids. The material from B was obtained with permission and in collaboration with Sanjeev Chawla in the Department of Radiology at the Perelman School of Medicine at the University of Pennsylvania.
A potential alternative is ultra-high-field 7T MR imaging used in conjunction with MRS.17 With substantially increased SNR and moderately improved spatial resolution, 7T has received US FDA and Conformité Européenne mark approval for clinical use.18 Comparisons of spectroscopy at lower field strengths such as 1.5T and 3T versus higher field strengths such as 7T to date have indicated that 7T is associated with substantial gains in SNR and more limited increases in spatial resolution,19 better spectral separation and therefore additional tumor-specific metabolite information,20 superior quantification of metabolites with small signals such as Gln,21 and reduced Cramér-Rao lower bound (CRLB) values.22⇓⇓-25 CRLB is a measure of the precision of an estimate, similar to an SD, which is used particularly with LCModel (http://www.lcmodel.com/). Despite evidence that these advantages of 7T may address the pitfalls of 3T MRS, its clinical usefulness has not yet been sufficiently explored.26,27 We aimed to investigate its utility by examining the literature published on 7T MRS in patients with HGGs. Most review articles published to date on ultra-high-field MR neuroimaging have had fairly broad scopes, highlighting techniques besides spectroscopy16,17,28,29 and/or neurologic disorders other than gliomas.15,26,30 A more targeted review of the use of 7T MRS to detect metabolites in HGGs is important to describe the current state of the field and provide clinical insight into new areas of investigation. In this article, we highlight the capacity of 7T to distinguish metabolic biomarkers. We also evaluate different protocols to determine optimal spectroscopy sequences. Finally, we turn to questions that remain unanswered. Given that the application of higher-field-strength MRS to glioma-related oncometabolism is still in its early stages, the full potential of this technique has yet to be realized.
Use of 7T MRSI to Aid in the Detection of Metabolic Markers
Resolution of Overlapping Resonances.
7T MRS is more sensitive to individual metabolites in HGGs.31 These include amino acids such as aspartate and lysine, which may be more difficult to visualize with MRS at conventional field strengths.31 At higher field strengths, MRS benefits from increased spectral separation. This helps to resolve overlapping resonances that occur at lower field strengths, making metabolic peaks, particularly those of coupled compounds, better separated and easier to quantify. By moderately reducing spectral overlap with better signal and chemical shift dispersion, 7T MRS may resolve specific metabolites more effectively than 3T on the basis of data from comparative studies on the 2 field strengths.21,23,25,32 Among these metabolites are Glu and Gln. Several studies comparing 3T versus 7T have found that improvements with 7T are particularly noticeable for Glu and Gln, which can be harder to individually measure at 3T due to substantial spectral overlap.22,33 The more precise quantification of these 2 metabolites has been primarily attributed to improved SNR and chemical shift dispersion at 7T.22 Compared with 3T, 7T also improves detection of 2HG, discriminating between 2HG and other metabolites that overlap on conventional 1D spectra, including Glu, Gln, and gamma-aminobutyric acid (GABA).33
A specific technique that has proved capable of detecting coupled metabolites and resolving overlapping resonances is 2D localized correlated spectroscopy (L-COSY) SVS, which separates and resolves metabolite signals along 2 different frequency dimensions.34 Although it is still largely in early exploratory stages, this technique has the potential to resolve complex resonances and separate phosphocholine (PC) from glycerophosphocholine (GPC), Lac from lipids, and Glu from Gln at 7T.34 2D L-COSY sequences at 7T result in clear identification and quantification of cross-peaks from metabolites such as GABA, Glu, Gln, glutathione (GSH), isoleucine, lysine, and compounds with Cho with increased separation compared with the same sequences at 1.5 or 3T.35⇓-37 In 1 study directly comparing one such 2D L-COSY sequence at 3T versus 7T (with the same scan times and voxel sizes at both field strengths), spectra generated at 7T had better signal quality than those at 3T.37 Cross-peaks to aspartate, GABA, GPC/PC, isoleucine, Lac, and mIns + Cho were readily apparent in 7T spectra but were weak or absent from 3T spectra.37
Providing greater spectral dispersion than traditional 1D spectroscopy, 2D L-COSY SVS has emerged as the higher-sensitivity technique.38 By indirectly recording T1 evolution, 2D L-COSY incorporates a second spectral dimension, detecting the transfer of coherence through cross-peaks between J-coupled metabolites. Metabolites with resonances that would ordinarily manifest as co-resonant with other metabolites become easier to separate. Nevertheless, 2D L-COSY also involves long acquisition times and sparse postprocessing support.38 Thus, shortening acquisition times and refining postprocessing techniques are vital. One group addressed these issues by constructing a nonuniformly weighted sampling scheme that reduced acquisition time by 25% and still preserved the same SNR, detecting diverse metabolites including GABA, Glu, Gln, GSH, Cho, and PC. These improvements, involving advanced spectral editing in 2D, make 7T MRS more clinically feasible by increasing the reliability of metabolic quantification.37
Separating peaks is crucial because it improves differentiation of individual metabolites, which have concentrations that are very small (typically expressed in mmol/l−1 or μmol/g−1 of examined tissue) and, therefore, difficult to detect.39 In spectroscopy, the strongest element of the signal is from water and is around 10,000-fold greater than the strongest metabolite signal from NAA.39 This separation of peaks is also possible at 3T, albeit with higher uncertainty. In addition, the resolution improvement due to this increased chemical shift dispersion at 7T may be diminished by the broadening of linewidths that occur at higher field strengths.39,40 However, optimized shimming and smaller voxel size can at least partially compensate for broader linewidths and still maintain some of the enhanced resolution associated with 7T.37,41⇓-43 Future work will further probe the capacity of 7T to separate metabolic peaks and will focus on other important factors such as uncertainty estimates and the amount of covariance.
Enhanced Characterization of Metabolic Pathways in Mutant IDH Gliomas.
The spectral quality of 7T may shed light on mutant IDH1 and IDH2 gliomas that produce the characteristic metabolite 2HG. The 2 mutant IDH1 and IDH2 enzymes participate in a reduction of α-ketoglutarate instead of the typical oxidation reaction, producing 2HG.44 Thus, α-ketoglutarate indicates an IDH-mutant profile. At higher field strengths such as 7T, which result in better spectral separation and more exact metabolic readings, 2HG forms a conspicuous signal even after a brief period of data acquisition.45 The improved spatial resolution, SNR, and spectral separation account for the sensitivity of 7T to this metabolite. 7T SVS has been shown to quantify 2HG with a high degree of specificity, making it easier to see whether this metabolite is present and ascertain the IDH status.27,33,34 One study detected 2HG in all 12 patients with IDH-mutant and/or radiography-suggested gliomas under investigation, with the mean concentration of 2HG measured at 3.1 (SD, 1.7) mM and a mean CRLB of 5% (SD, 2%).46 In one of these patients with an IDH1-mutated oligoastrocytoma, the 2HG was estimated to be 6.2 mM, with high precision (CRLB = 2%).46 Four patients had tumors with lower 2HG concentrations (<2 mM), which were still acquired with acceptable precision (CRLB ≤7%).46 Another recent study with 7T SVS estimated 2HG to be within the range of 2.3–3.3 mM in tumors of patients with mutant IDH gliomas.45 Thus, 2HG is an indicator of tumor profile and evolution.46 7T SVS has also revealed associations between 2HG and mIns, glucose, and taurine.47
Although not every spectroscopy sequence readily quantifies 2HG, the ease with which certain 7T MRS sequences single out this oncometabolite might provide more insight into survival prospects. Optimizing MRS measurements of 2HG at 7T is, therefore, critical. The optimal TE of the point-resolved spectroscopy sequence (PRESS) at 7T was shown to be 78 ms (TE1 = 58 ms, TE2 = 20 ms) by Ganji et al,46 who used density-matrix simulations and phantom validation to enhance signal selectivity for 2HG. Notably, this PRESS TE was superior to short-TE MRS, allowing clearer differentiation between 2HG and other metabolites including Glu, Gln, and GABA.46 Quantifying 2HG is more difficult at lower field strengths because this oncometabolite often overlaps with Glu, Gln, GABA, and NAA.48 However, it is still possible to quantify 2HG at 3T if specific strategies are used.49,50 These include using the optimal PRESS TE of 97 ms established by Choi et al.51,52 Other potentially useful approaches at 3T involve spectral editing and the 2D correlation method53 and hyperpolarized 13C MRS.54 Another strategy for 3D imaging of 2HG at 3T involves echo-planar spectroscopic imaging with dual-readout alternated gradients, which 1 recent study used to generate images of 2HG with a high level of precision (CRLB, <10%).55
Nevertheless, 2HG substantially benefits from the greater SNR and spectral separation attainable at 7T, as supported by the results from a study by Ganji et al46 that involved output results from LCModel. Serving as a prior knowledge-based spectral fitting model of complete spectra instead of singular peaks, LCModel evaluates an in vivo spectrum as a linear combination of in vitro spectra from individual metabolite solutions.56 LCModel, thereby, enables the resolution of spectra that are virtually identical in 1 frequency area, provided that they have distinct signals at other points in the spectrum. In this study, although mean estimates of 2HG and total choline (tCho) were not meaningfully different at 7T and 3T, the mean 2HG CRLB was markedly lower at 7T (5%) than at 3T (8%) due to the higher SNR and spectral resolution associated with higher field strengths.46 In addition, the mutual dependence of 2HG and GABA signal estimation was much smaller at 7T than at 3T.46 At 7T, 2HG and GABA signals had substantial opposite polarity and apparent narrowing.46 Thus, this improved signal separation may permit more facile discrimination of 2HG.46 This long TE of 78 ms, which is specific to 2HG, can be expanded to 2D MRS at 7T with appropriate modifications. For example, 1 study performed prescription of a VOI based on the 7T 2HG-optimized PRESS sequence before echo-planar spectroscopic imaging with dual-readout alternated gradients.45 Thus, the same TE parameters can be applied in different spectroscopic contexts at higher field strengths.
Beyond measuring 2HG, 7T SVS has proved capable of detecting minute differences between IDH1 and IDH2. Mutant IDH2 gliomas have higher 2HG than IDH1 gliomas, with more elevated 2HG:Cho and 2HG:Cr in IDH2 gliomas than in IDH1 gliomas detected in 2 recent 7T SVS studies.27,47 In 1 recent 7T SVS study, mutant IDH2 gliomas also had higher mIns and Cho compared with IDH1-mutant gliomas.47 Thus, 7T MRS may strengthen visualization of metabolic differences between mutant IDH1 and IDH2 gliomas.47 These findings may lead to precision medicine tailored to patients’ IDH statuses.47 Distinguishing IDH-mutant and IDH wild-type tumors is important because mutant IDH1 and IDH2 gliomas have better prognoses.57 A prospective analysis found that patients with grade II, III, and IV IDH-mutant gliomas survived longer than those who had IDH wild-type.51,52 Other studies have found similarly robust associations between the presence of IDH mutations and increased survival.58⇓-60A potential explanation may be that IDH responds better to targeted treatment.57,61 Thus, IDH mutations have prognostic value, predicting more favorable responses to radiation and chemotherapy.57,61 The fact that these mutations are common in grade II and III gliomas and in secondary GBM makes them even more appealing.
The potential advantages of using 7T MRS to spot this oncometabolite extend beyond survival time to treatment. Several inhibitors of mutant IDH have already been engineered.62 7T MRS may complement this therapeutic targeting by zeroing in on IDH-mutant gliomas and further differentiating between IDH1 and IDH2 mutants.27,47 These 2 enzymes have unique characteristics that can be independently targeted by different drugs with distinct mechanisms. These include their location in cells, with IDH1 typically found in the cytosol, and IDH2, in the mitochondrial matrix.63 7T MRS could, therefore, help gauge whether a patient has an IDH-mutant glioma and, if so, whether the glioma is IDH1 or IDH2, dictating the most appropriate therapy.27,47 Application of this technique would, in turn, direct novel molecular therapies that impede IDH-mutant tumors and thereby prolong survival.64,65 During the past year, IDH has gained increasing recognition as an important tumor marker in the WHO’s 2021 Classification of Tumors, which called for grouping all IDH-mutant diffuse astrocytic tumors into 1 category based on IDH status (astrocytoma, IDH-mutant) and subsequently grading them as CNS WHO grade II, III, or IV.66 Given that IDH has therefore become even more central in defining gliomas, using techniques such as 7T MRS to better visualize this oncometabolite, a potential personalized biomarker,47 may be worthwhile. Most important, immunohistochemical and molecular pathologic analyses of resected tissue acquired through surgery remain the criterion standard for the diagnosis of IDH-mutant gliomas.27 However, there is growing interest in noninvasive approaches, including imaging, as a complement to conventional invasive techniques.67⇓-69 To that end, it is worth considering whether 7T MRS might be one such noninvasive tool that can be used to gather additional information about IDH-mutant brain tumors, which may inform tumor diagnoses, survival predictions, and treatment strategies.47 Specifically, noninvasive quantification of 2HG levels with MRS may be used to track targeted therapy and guide decisions about possible changes in therapy when necessary.34 To that end, MRS may have added value when used in conjunction with traditional immunohistochemistry following biopsy or surgery.34
Broadening of Possible Therapeutic Targets: Glu, Gln, and Gly.
7T MRS enhances visualization of other metabolites that may serve as therapeutic targets. Glu has been implicated in HGG metabolism and can be identified with 7T MRS, which is capable of isolating Glu from Gln so that these 2 metabolites can be evaluated separately (Fig 2).70 Separating Glu from Gln is often challenging at 3T with combined Glu and Gln denoted as Glx, but it is easier at 7T, which can differentiate the 2 metabolites, albeit with variable spatial resolution quality.31 Distinguishing Glu from Gln allows deeper insight into metabolic pathways in gliomas and is particularly clinically relevant given the growing evidence for the influence of glutamatergic synapses on tumor progression.71 Glioma cells have been shown to secrete Glu, precipitating a rise in excitotoxic, extracellular Glu and promoting malignant growth.72 Thus, Glu plays a critical-but-complex role in tumors that 7T MRS may help to better characterize.
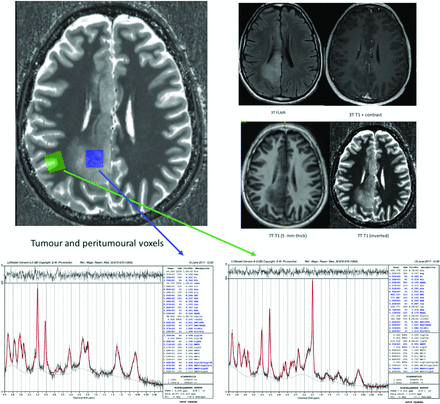
3T/7T MR imaging data, 7T SVS data, and LCModels from a patient with diffuse glioma are presented (TR/TE for the 7T SVS data = 8500/6 ms), with tumor data on the left (blue arrow) and peritumoral data on the right (green arrow) for each patient. Inspection of the MRS metabolic profiles shows that the peritumoral region has a relatively normal profile except for a highly elevated Glu concentration compared with both Gln and Cr. This is in contrast to the tumoral region, where there is a complete reversal of this profile with Gln and Cho being elevated compared with Glu, NAA, and Cr. These data provide further evidence of the dysregulation of the Glu-Gln shuffle, serving as a source of seizures in patients with gliomas. This material was obtained with permission and in collaboration with Andrew Neal and Bradford A. Moffatt at the Melbourne Brain Centre Imaging Unit (MBCIU), the University of Melbourne node of the Australian National Imaging Facility.
In a recent study evaluating 23 HGGs with 7T MRSI, increased Glu was found in IDH wild-type tumors, while decreased Glu was found in IDH-mutant tumors70 Although additional studies with larger sample sizes and low-grade gliomas (LGGs) are necessary to validate these preliminary findings, this trend suggests that Glu could be a potential marker for the absence of IDH mutations.70 Another 7T MRSI study found that the ratio of Glu/total Cr was substantially decreased in grade II gliomas compared with normal-appearing white and gray matter.31 Widespread reductions in Glu were also observed in 9 grade II, III, and IV gliomas in a separate study involving 7T MRSI.73 Considering its role in glioma proliferation and survival as both a metabolic intermediate and the primary excitatory neurotransmitter in the brain, Glu is ripe for targeted interventions.74 Yet despite the essential part Glu plays in tumorigenic processes, few other studies have used 7T MRS to investigate the spatial profile of Glu in patients with gliomas or to rigorously evaluate whether 7T MRS might be able to identify differences in Glu levels in different grades of gliomas.70 Observing how Glu varies in larger subject populations could refine the differential diagnoses of LGGs and HGGs.
Gln is likewise altered in gliomas.75 Changes in Gln are more appreciable at higher field strengths.73 Increased Gln has been detected in a range of gliomas.31,70,73 Serving as a substitute for glucose in the tricarboxylic acid cycle, a regulator of oncogene expression and a suppressor of apoptosis, Gln may be an imaging marker for tumor cells, helping to distinguish gliomas from other neurologic disorders.76 A recent study with 7T MRSI discovered elevated Gln in HGGs, including oligodendrogliomas, diffuse and anaplastic astrocytomas, and GBMs.73 In a patients with a grade IV GBM, increased Gln overlapped with contrast-enhanced T1WI regions, decreased NAA, and widely reduced metabolic activity in the center of the tumor. Widespread increases in Gln were also observed in a similar 7T MRSI study, further supporting the centrality of Gln in tumor metabolism and homeostasis.31 Other 7T studies have also yielded results suggesting that Gln might be a potential tumor biomarker, with one reporting that Gln was almost 100% higher in tumor tissue.71,77
Gln is present in several different HGGs and could, therefore, be a target for antineoplastic drugs used to treat many patients with gliomas.78 In the first step of glutaminolysis, glutaminase catalyzes the deamidation of Gln to Glu, which then serves as a substrate in the tricarboxylic acid cycle or contributes to GSH synthesis (Fig 3).79 Glutaminase inhibitors, which prevent the conversion of Gln to Glu and thereby decrease tumorigenesis, have the potential to treat HGGs with high Gln levels.80 Increased Gln could, therefore, be an indicator of treatment with glutaminase inhibitors. 7T MRS could confirm the status of Gln as a biomarker for HGGs, resulting in more targeted therapies that attack Gln pathways.81 Suppressing these pathways might be effective because Gln supports the growth of highly proliferative cells, especially those in HGGs. Gln contributes to anaplerotic reactions that sustain glycolysis and give malignant tissue an advantage over normal tissue.81 Gln also provides building blocks for nucleotide synthesis and for the vital antioxidant GSH, both of which make gliomas more resistant to radiation and chemotherapy.82 Glutaminase inhibitors that exploit the addiction of the cancer cells to Gln would curtail the development of HGGs. In the past few years, CB-839 has been the primary glutaminase inhibitor under investigation for the treatment of multiple cancers in clinical trials, most of which have been Phase I and II.83⇓-85 A recent pharmacometabolic analysis showed that CB-839 effectively inhibited glutaminase activity and curtailed the growth of glioma stemlike cells with high target specificity.79 These glioma stemlike cells lead to drug resistance and tumor recurrence in GBM.79 Given that it quantifies levels of Gln so precisely, 7T MRS could reshape glioma treatment, tipping the balance toward more personalized treatments with these glutaminase inhibitors.81
Major glutamine pathways in HGGs are featured. Gln undergoes a deamidation to Glu, which subsequently promotes tumorigenic growth either as a substrate in the tricarboxylic acid (TCA) cycle or as a precursor to GSH. The deamidation reaction is catalyzed by glutaminase. This enzyme is the target of glutaminase inhibitors, which may be useful therapeutic agents for patients with gliomas that involve high levels of glutamine detected by 7T MRS. PEP indicates phosphoenolpyruvate.
Nonetheless, at present, there are very few published 7T MRS studies investigating the significance or clinical utility of Gln measurements in HGGs. As previously discussed, Gln has long been an established precursor of both Glu and GSH and is known to contribute to the cellular energy process and lipid synthesis, suppress apoptosis, and potentially protect cancer cells from radiation therapy.31,77 In view of its major role in tumor metabolism, biosynthesis, and homeostasis, it would certainly be worth investigating whether the abundant Gln detected in HGGs via 7T MRS may be clinically relevant, serving as a biomarker or therapeutic target.70,73 More rigorous evaluation of Gln at 7T and even at lower field strengths will provide further insight into how quantification of this metabolite might aid in the management of these tumors.
Gly has long been suspected of being an imaging biomarker for brain tumors, with increased Gly in HGGs including GBMs.86⇓-88 One of the major challenges to date has been separating Gly from mIns. The differentiation between mIns and Gly has been accomplished at 3T with a long-TE in other individuals with other disorders, including alcohol use disorder89 and nonketotic hyperglycemia.90 Nonetheless, studies with spectroscopy at 1.5T and 3T have not separated Gly from mIns in HGGs. Although it is still difficult to differentiate these 2 metabolites on the basis of their chemical shifts even at higher field strengths, there is emerging evidence that 7T MRSI may be better equipped to isolate Gly70 (Fig 4). Gly may aid in tumor proliferation by serving as an intermediary in nucleotide and Glu synthesis and could, therefore, be a promising metabolic metric.91 In a recent study on 3T MRSI, every tumor with increased Gly had postcontrast enhancement, a high cell proliferation rate (MIB-1), and shorter survival.91 The correlation between Gly and MIB-1 indicates that HGGs may remodel Gly metabolism to sustain rapid cell proliferation. These findings support the clinical importance of Gly, with higher levels of Gly correlating with poorer clinical outcomes in patients with HGGs.
Exemplary 7T concentric ring trajectory FID-MRSI spectra of a patient with GBM. This method uses FID acquisition combined with concentric ring trajectories, bringing together multiple potential benefits: high resolution and acceleration while maintaining sufficient SNR, low SAR, B1 insensitivity, no selection box, and detection of J-coupled metabolites with 7T spectral separation. This measurement was acquired in 15 minutes with a nominal isotropic resolution of 3.4 mm, covering the cerebrum. Well-defined metabolic peaks are observed at 3 points: a tCho and Gly hotspot, a tCho and Gly cold spot, and normal-appearing white matter (NAWM). It is noticeable that even in the tumor tCho cold spot, tCho concentrations are higher than in NAWM but about 50% lower than in the tCho hotspot. Two of the metabolites that appear in these plots (Gly and Gln) have emerged as promising therapeutic targets and may serve as imaging biomarkers for patients with HGG. This material was obtained with permission from Gilbert Hangel at the High-field MR Center at the Medical University of Vienna. T1w indicates T1WI; tCr, total Cr.
Gly could help clinicians differentiate glioma grades, adapt treatment plans accordingly, and predict survival time on the basis of the levels of Gly visualized with 7T MRS.91 Predicting survival is especially important for HGGs, given that patients with grades III and IV gliomas have worse chances of survival than those with LGGs.92 However, while several studies have used lower field strength MRS to probe the prognosticative power of Gly, few have explored whether 7T MRS might further validate Gly as an index of HGG aggressiveness. Given that it may be more able to separate out Gly than spectroscopy at lower field strengths, 7T MRS might be able to address questions about whether this metabolite can indeed mark the progression from low- to high-grade disease and from posttreatment to recurrent disease, fully establishing Gly as a biomarker and improving patient care.91
Optimization of 7T MRS Sequences
Because 7T MRS is still novel, there is not a criterion standard MRS pulse sequence. The fact that different sequences capture distinct metabolites depending on the clinical context has made it challenging to reach a consensus. A quandary still unresolved is the ideal TE. Transitioning to longer TEs enables differentiation of 2HG from adjacent GABA and Glu.46 Nevertheless, short-echo 3D MRSI also differentiates Glu and Gln.93 Future studies will elucidate the optimal TE length. Nonetheless, there are still common threads among successful protocols. Balancing spatial resolution and measurement time is crucial, as is optimizing the SNR. Rapid concentric ring trajectory encoding was recently shown to shorten TRs and enhance SNR efficiency.77 Dual-readout alternate gradients used in echo-planar MRSI at 7T have also been found to have similar effects on SNR efficiency, resulting in high-resolution imaging of metabolites such as 2HG.45 The TE for any given acquisition will depend on the precise variables of interest for a particular patient, and >1 acquisition with different TEs may be required, as is often the case for clinical spectroscopy. Thus, these may be additional strategies to consider.
Techniques to augment SNR allow improved resolution and smaller voxel volumes, which consequently enable better spatial localization of differences among individual gliomas.73 These could, in turn, help define tumor margins, enhancing visualization of metabolic abnormalities. A method to improve the SNR is to use high-channel-count radiofrequency (RF) receive array coils, which have been shown to provide better SNR than volume coils, especially near the cortical surface.94 Furthermore, new coil technology can improve spectral linewidth by incorporating B0 shim capabilities in the form of spherical harmonic shim insert coils or local multicoil shim arrays.41,42 In 1 realization of the local multicoil shimming approach known as the AC/DC coil, independent DC currents are driven in RF receive loops to provide many dfs for high-spatial-order B0 shimming of the brain. This tool was recently shown to improve metabolite linewidth and refine quantification, allowing more brain coverage and the detection of 2HG at higher resolution (Fig 5).43
7T MRSI with standard second order shimming and higher order shimming with an AC/DC shim array was used to visualize Cho and 2HG in an IDH1 astrocytoma. A metabolic hallmark of IDH1 and IDH2 gliomas, 2HG generates a particularly strong signal at 7T, which may make it easier to diagnose patients with IDH-mutant profiles. The SNR of 2HG is lower in this example because this patient was treated with an operation and radiochemotherapy, which decrease 2HG levels. The primary clinical objective in this study was to determine whether there were residual mutant IDH tumor cells posttreatment. The white arrows indicate the frontal area where there are missing voxels in the Cho metabolic map and falsely increased values in the 2HG map due to a larger spectral linewidth obtained with second order shimming. The tumor has a higher contrast-to-noise ratio in the 2HG and Cho maps obtained with the AC/DC shim array. Examples of spectra are shown from tumor and healthy brain in the frontal region (corresponding to the white arrows). The position of the 2HG peak at 2.25 ppm is indicated in all spectra. For the adiabatic spin-echo excitation with TE = 78 ms, a negative peak should be obtained for 2HG, which is clearly visible in the tumor spectrum with AC/DC shim but not apparent in the tumor spectrum with standard second order shimming (2SH) shim. In particular, the frontal spectrum obtained with the 2SH shim is completely destroyed by the B0 inhomogeneity, while metabolite peaks are clearly visible with the AC/DC shim. 2HG is falsely fit in the frontal healthy spectrum with the 2SH shim due to negative spectral artifacts that appear at the 2.25-ppm 2HG peak position. The dashed vertical line indicates the location of the main 2HG peak at 2.25 ppm, which should be negative at 7T and TE = 78 ms. All the voxels in the MRSI were fit to create a metabolic map. A spectrum from the frontal voxel was chosen to show a false-positive fit of 2HG in healthy brain when the spectral quality is not adequate. Frontal brain regions are difficult to shim with standard methods, and the AC/DC coil can improve B0 homogeneity due to additional B0 shimming. Frontal loops in AC/DC are very close to frontal brain areas and create complex B0 field patterns to shim out the susceptibility induced by air tissue around the frontal sinus. The second order shimming was obtained with the manufacture’s software. This material was obtained with permission from and in collaboration with Jason P. Stockmann and Ovidiu C. Andronesi at the A. A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital and Harvard Medical School. LW indicates linewidth.
Transceiver arrays also address the increased B1 inhomogeneity and RF power deposition typical of 7T MRS. These 2 issues, along with acute chemical shift localization (CSL) errors, can result in image artifacts and reduced spatial coverage.95 Chemical shift localization errors are the spatial shifts in the excited volume for metabolites that resonate at varied frequencies.95 In 1 study, using an 8-channel transceiver array minimized these problems, harnessing the power of 7T to visualize metabolites such as Glu.93 Another way to mitigate B1 inhomogeneity involves adiabatic pulses, a special class of RF pulses that can be integrated into existing MR imaging pulse sequence structures to provide more B1-insensitive excitation and refocusing of magnetization.96 However, while these adiabatic pulses yield a more uniform B1 profile, they also result in more RF deposition, which may have clinical implications if measures are not taken to remain below the specific absorption rate (SAR) safety limits set by the FDA and the International Electrotechnical Commission.95,97
Considering that SAR, a quantitative measure of RF power deposition, also increases with field strength, conducting spectroscopy at 7T poses unique problems.98 Balancing the increased RF power associated with higher field strengths and concerns about patient safety has limited the clinical translation of 7T MRS so far. Nevertheless, a recent study demonstrated that using an adiabatic Shinnar Le-Roux algorithm helps generate a fully adiabatic envelope and a more homogeneous distribution of RF, bypassing some of these obstacles.95 The pulses obtained with these adiabatic 180° RF pulses, part of a Semi-Adiabatic Spectral-spatial Spectroscopic Imaging (SASSI) pulse sequence, quantified major metabolites while alleviating B1 heterogeneity, staying within the range of prescribed RF limits and attaining an SNR similar to that in the leading adiabatic alternative, semi-LASER, with just a third of the SAR.95 The increased B1 insensitivity and reduced chemical shift localization errors suggest that SASSI might be even more powerful than the aforementioned semi-LASER. Incorporating pulse sequences such as SASSI, which has a more uniform B1 profile and lower SAR, might, therefore, be warranted. Another approach might be to use a free induction decay (FID) acquisition, which bypasses high SAR pulses altogether. This acquisition strategy minimizes SAR by avoiding multipulse schemes, reducing T2*-decay even below short-TE approaches, and limiting the signal evolution of J-coupled metabolites such as Gln, Glu, mIns, and taurine.70
Both susceptibility artifacts and B0 inhomogeneity, directly proportionate to field strength, also detract from 7T MRS and skew the geometry and intensity of images. The variations between voxels appear as spectral shifts for metabolite peaks, making frequency-selective pulses that depend on specific spectral bands less accurate and methods for suppressing water and lipid signals more complicated.99,100 As a result, metabolic peaks widen, peak overlap increases, and SNR decreases. The best way to avoid these obstacles is to use strong referencing schemes, sophisticated B0 shimming approaches such as dynamic multicoil shimming, and reduced voxel volumes, all of which help to increase SNR and spectral separation.30,101
The incorporation of techniques such as compressed sensing might also benefit 7T MRS. Dependent on nonuniform k-space undersampling and the assumption of spatial and/or spectral sparsity, compressed sensing MR imaging was first combined with 1H-MRSI in 2009 in vitro102 and in 2012 in vivo.102,103 Compressed sensing 1H-MRSI has been used for brain imaging in conjunction with PRESS,103 semi-LASER prelocalization,104 and section-selective 1H-FID-MRSI.105 There are several challenges associated with combining compressed sensing with 1H-MRSI. Large water and lipid nuisance signals can prevent reconstruction algorithms from sensing lower-intensity metabolite peaks due to misadjusted thresholding.106 The fact that 1H spectra, including MRS with a short-TE, are not sparse can also complicate acceleration along the spectral dimension.106 However, if these obstacles can be overcome, the application of compressed sensing to MRS has the potential to improve spatial resolution with only minimal increases in acquisition time. Most work to date has involved compressed sensing in spectroscopy at lower field strengths such as 3T, and very few studies have used compressed sensing at higher field strengths such as 7T in investigations of the human brain. However, a very recent study used an effective acquisition-reconstruction scheme involving a 1H-FID-MRSI sequence, a short-TR acquisition, compressed sensing acceleration, and low-rank modeling with total generalized variation constraint for metabolic imaging in 2 and 3 dimensions at 7T in 5 healthy subjects, ultimately generating images with distributions that were highly specific to individual metabolites.107 Given the high sensitivity and short acquisition time achieved by this group, expanding the use of compressed sensing in 7T MRS would be worthwhile, taking advantage of the additional SNR and other potentials of this higher field strength.
Expanding Horizons: Improved Detection of Infiltrated Tissue and Treatment Effect
Infiltrated Tissue.
As a result of its sensitivity to metabolites ranging from Glu to 2HG, MRS offers another way to localize diffuse glioma infiltration that can complement structural information provided by conventional MR imaging.108,109 A metabolite detected by MRS that has emerged as a strong indicator of the range of tumor infiltration is Cho (Fig 6).110 The boundaries of both HGGs and LGGs as delineated by MRSI on the basis of Cho-containing compounds correlate with histopathologic analyses, further validating the capacity of spectroscopy to assess the degree of tumor infiltration.110 Thus, mapping Cho with MRS may help demarcate tumors in treatment planning. More extensive infiltration would be a signal for more aggressive surgical resection or radiation therapy to prevent recurrence.111 Modifying radiation therapy to account for infiltrated tissue could improve patient outcomes. Increasing high-dose radiation therapy boost volumes, for instance, could decrease the likelihood of recurrence and increase survival rates.112
T1-weighted contrast-enhanced MR imaging (left) of a patient with a glioma subsequently diagnosed with recurrent tumor (true progression) and the same image with a superimposed map of the Cho/NAA ratio (right) acquired using echo-planar spectroscopic imaging. The map shows a hotspot of the elevated Cho/NAA ratio in a region that does not show contrast uptake on postcontrast T1 MR imaging (red depicts Cho/NAA > 1).
MRS could, therefore, help to improve HGG treatment strategies by capturing tumor expansion via metabolic mapping. The limited spatial resolution and spectral sensitivity of MRS at lower field strengths has, heretofore, impeded the usefulness of this technique in detecting tumor infiltration. Given that 7T MRS allows higher spatial resolution, increased SNR, and improved spectral separation, with the highest in-plane resolution published at 7T involving an MRSI ultra-high-resolution sequence with a 128 × 128 matrix and a nominal voxel volume of 1.7 × 1.7 × 8 mm3, the question emerges: Might performing spectroscopic imaging at higher field strengths address these concerns and refine approaches to diagnosing and treating HGGs?.113 Future work will clarify whether 7T MRS might provide even more insight into infiltration than spectroscopy at lower field strengths.114⇓-116
Treatment Effect.
MRS may also strengthen visualization of pseudoprogression, a treatment effect described by the Response Assessment in Neuro-Oncology criteria, in which chemoradiotherapy-induced lesions imitate tumor progression but ultimately stabilize or improve with time.117⇓-119 Seen in 20% of patients who have undergone treatment, these alterations are produced by temozolomide, which has become the standard of care for GBM.120 Although they closely resemble more malignant pathology, these changes do not reflect actual tumor growth.121 In contrast to tumor cells, most of these modifications are asymptomatic.120 In fact, many even resolve months after patients have finished temozolomide.122-123 While these lesions are not clinically worrisome, they have complicated neuroradiologic studies.124 This treatment-related change is radiographically similar to tumorigenic growth.125 Both exhibit nearly identical contrast-enhancement patterns on conventional MR imaging, which does not differentiate pseudoprogression from progressive disease.126
The dearth of imaging parameters that reliably identify these lesions complicates decisions about how to best manage patient care because the approaches to dealing with pseudoprogression versus true progression are radically different.127 Whereas patients in the latter group require further treatment for persistent tumorigenic activity, those in the former group have a better chance at spontaneous recovery.128 To prevent patients with pseudoprogression from undergoing avoidable surgery or chemotherapy, seeking noninvasive ways to distinguish pseudoprogression from true progression is essential.129-130 Using techniques such as MRS is a possible solution.131 The metabolic details contained within spectroscopic images may help distinguish largely innocuous disease from recurrent glioma, ushering in tailored treatment strategies.132 Although differentiating pseudoprogression and true tumor progression is still difficult even with MRS, this technique has shown the potential to reveal differences in the spectroscopic features of malignant tissue and treatment effect–damaged tissue.121,127,133,134 Whereas recurring tumors often have increased Cho due to higher cell membrane turnover, therapy-associated lesions frequently have slightly decreased NAA and substantially decreased lipid-Lac peaks.121,127,135,136
In a recent meta-analysis of advanced MR imaging techniques, SVS had the highest diagnostic accuracy, reliably discerning treatment-related changes in patients with HGG.137 Specific parameters in SVS may be highly sensitive to metabolic differences.138 These include the ratios of Cho/Cr and Cho/NAA, with higher values in recurring tumors.139⇓-141 A study with 2D chemical shift imaging SVS at 1.5T found that these ratios were 97% accurate in distinguishing pseudoprogression and tumor advancement.142 Another study with 3T SVS involving the same 2 Cho metrics reported a similarly high level of accuracy, with 94.1% sensitivity and 100% specificity.143 As encouraging as findings such as these are, the differential diagnostic utility of MRS continues to be hampered. A long-standing roadblock has been the metabolic overlap between tissue treated with temozolomide and tissue containing actively growing malignant glioma cells on spectroscopy at 1.5T and 3T.121 Moreover, there is not yet any definitive agreement as to which spectroscopic parameters and threshold levels can best differentiate pseudoprogression from true progression.118 Both types of lesions also have similar metabolic fingerprints, including low NAA and high Cho, Lac, and lipids.121
The high similarity demands more nuanced spectroscopy that can disentangle the 2 on the basis of subtle discrepancies.144 Once again, 7T MRS surfaces as a prime candidate, well poised to sort out problematic overlap and pinpoint minute metabolic differences. Potential metabolic candidates include Glu, Gln, and Gly. Yet despite all these attributes, no studies have analyzed the diagnostic accuracy of 7T MRS in distinguishing pseudoprogression. In view of its sensitivity and specificity, there is no doubt that this system would be worth exploring. If 7T MRS does provide greater sensitivity to these metabolic variations in proportion to the expectedly higher SNR, then such a discovery would take analyses of treatment effects and therapeutic tactics in new directions.145 With 7T MRS, differentiation between pseudoprogression and authentic progression might finally become a reality, improving predictions about prognoses and directing calls about therapy.146
3T versus 7T: Qualitative and Quantitative Differences
A summary of the qualitative strengths and weaknesses of 3T and 7T is included (Table). In terms of quantitative metrics, as discussed throughout this review, one of the major benefits of 7T is higher SNR, which increases substantially and possibly even supralinearly147 with B0, resulting in modestly enhanced spatial resolution and contrast.26 Coil design can further elevate SNR at higher fields, leading to a 2- to 6-fold increase under certain conditions.26,148 A study investigating SVS at 3T and 7T using 32-channel head coils found that mean SNR values at 7T were substantially higher than at 3T (135 [SD, 28], 116 [SD, 33], and 138 [SD, 29] versus 83 [SD, 12], 97 [SD, 7], and 83 [SD, 5]) in brain regions under examination (the anterior cingulate cortex, centrum semiovale, and dorsolateral prefrontal cortex).22 Another study that also compared 3T and 7T recorded a 1.7-fold increase in the average SNR per unit of time at 7T, providing further support for the improved sensitivity at higher field strengths.23
Comparison of 3T versus 7T MRS in visualization of metabolic markers in HGGsa
Studies involving 7T have reported increases in linewidths of metabolites such as NAA, Cr, and Cho when measured in hertz but decreases when measured in parts per million.19 Thus, linewidths increase less than proportionally to field strength and are, therefore, higher in absolute terms in hertz but lower in relative terms in parts per million. Although the short-T2 relaxation times and susceptibility effects at high field contribute to an increased linewidth,32 the spectral resolution could remain increased at 7T with optimized shimming methods and smaller voxel volumes.37 Thus, proper compensation with techniques such as shimming is important to minimize broader linewidths that occur at higher field strengths, which can exacerbate spectral overlap and complicate the separation of metabolites, including Glu and Gln.149
The increased SNR associated with 7T MRS may be particularly helpful in the detection of Glu and Gln.149 These 2 metabolites can be reliably differentiated from one another with sophisticated techniques in multiple areas of the brain even at 3T.149 However, especially at such lower field strengths, Glu and Gln are frequently grouped together as a sum Glx (Glu + Gln), recorded in spectroscopic VOIs in neurologic regions with low SNR and broad linewidths.149 Combining these 2 metabolites into 1 discrete peak makes discriminating between them very challenging at 3T, particularly in certain areas of the brain.149 In contrast, at higher field strengths such as 7T, separating Glu from Gln is more achievable due to the potential for higher SNR and greater spectral separation.24
CRLBs have been shown to decrease as B0 increases for many metabolites under investigation in several studies, contributed by both the increased SNR and spectral resolution.19,150 7T resulted in higher measurement precision for aspartate (average CRLB 27% at 3T versus 15% at 7T), Glu (average CRLB 5% at 3T versus 3% at 7T), and Gln (average CRLB 19% at 3T versus 9% at 7T) in the 32-channel head coil study.22 Nevertheless, these differences in measurement precision were less appreciable for larger signals in the spectrum, with CRLBs for 3T and 7T very similar for metabolites including total NAA, total CR, tCho, and mIns and Gly.22 The benefits provided by 7T in terms of measurement precision (as quantified via CRLBs) may, therefore, vary depending on the specific metabolite of interest. However, there is growing evidence that 7T correlates with decreased CRLBs relative to 3T. Other studies have likewise discovered that Glu has a lower CRLB at 7T than at 3T, including one that involved 7T short-TE MRSI.25 Another study found that the CRLB averaged over 18 total metabolites was significantly lower (35% lower) at 7T than at 3T (P < .015).23 The CRLBs of Gln, τ, and Glx were the most visibly reduced by 7T.23 In addition, 7T measured Lac with CRLB <20% (final mean Lac concentration = 0.7 [SD, 0.1] mmol/kg, mean CRLB = 9% [SD, 1.6%], which was a distinct improvement compared with 3T.23 Yet another study similarly reported lower CRLB (P < .001) at 7T versus 3T for Glx in parts of the brain such as the thalamus and pons.24 At 7T, CRLBs of total NAA, total Cr, and mIns were also lower than at 3T.24
Given that 7T MRS has not yet been fully integrated in a clinical setting, there are not yet many statistics of clinical assessment, including tumor grading, available. Thus, comparing 7T with 3T in this area is difficult. Although studies have assigned grades II, III, and IV to lesions via histopathologic analysis before imaging with 7T and 3T,25 very few have conducted rigorous comparisons of tumor grading at these 2 field strengths and assessed correlations between 7T MRS imaging results and tumor grading. Most studies have concentrated on the capacity of 3T MRS to assess tumor grade.151⇓⇓⇓⇓⇓-157 One study found that Cho/Cr and combined Lipid-Lac/Cr ratios were helpful in differentiating LGGs from HGGs, with diagnostic accuracy higher (85.7% versus 82.9%) at short-TE or combined short-TE and long-TE than at intermediate-TE only.152 Another similarly reported that Cho/Cr, in conjunction with other imaging parameters, was an effective marker for distinguishing LGGs and HGGs with 87.0% sensitivity and 88.9% specificity.153 The usefulness of Cho/Cr in stratifying gliomas of different grades has been documented in several other 3T studies.155,156 Maximum Cho also proved in 1 study to yield even higher diagnostic accuracy (82.5% versus 72.1% accuracy) than Cho/Cr.155 In addition, Cho/NAA may ???????? in the differentiation of gliomas, with higher absolute values of Cho/NAA in HGGs than in LGGs seen in 1 study, which combined Cho/NAA with structural MR to achieve 86% sensitivity and 80% specificity.157
As the use of 7T MRS becomes more widespread in clinical contexts, similar appraisals of its ability to determine differences between gliomas and aid in the grading process should be undertaken. Although further study is warranted, the use of 7T MRS for tumor grading may have several potential benefits. Given the technical assets of spectroscopy at higher field strengths, including reduced CRLBs and modest improvements in spatial resolution, 7T MRS could further isolate Cho/Cr and Cho/NAA and could provide more information about other metabolites in different HGGs, thereby making the process of differentiating tumor grades less challenging. In particular, elevated Cho, reduced NAA, and levels of lipids and Lacs have been shown to correlate with higher tumor grade and aggressiveness.158 There are emerging signs that 7T may have a higher capacity than lower field strengths with poorer signals to reveal these kinds of differences in metabolite levels in cancerous-versus-normal tissue and in different gliomas, which may result in more refined detection and classification of tumors.20,31 However, additional investigations involving spectroscopy and tumor grading are necessary to confirm whether 7T is indeed superior to 3T in this regard.
Future Clinical Directions
More sensitive than conventional MRS to metabolic markers at lower fields, 7T MRS can detect and differentiate among HGGs, revealing different metabolic characteristics (Fig 7). The abnormal metabolic activity visualized by 7T MRS may advance disease etiology, diagnostic approaches, and therapeutic interventions.112 Nevertheless, 7T MRS of HGGs is still in its infancy. The lack of spectroscopic imaging studies with 7T indicates that advances must still be made.93 One of the most common problems in every report reviewed is the small sample size. Future studies will include larger groups of patients. Broadening the patient population will increase the statistical power and expand the 7T MRS database.70 Increasing the sample size will also provide information about metabolic heterogeneity in diverse HGGs.34 Furthermore, deeper insight into tumor microenvironments will elucidate metabolic signatures of various gliomas.159
7T MRSI can capture gliomas in different stages, such as the recurrent grade II oligodendroglioma (progression) in the newly diagnosed, presymptomatic patient in A and the recurrent grade II oligodendroglioma (stable) in a treated patient in B. These spectra were acquired at 7T and show differences between progressing and nonprogressing tumors. The progressing tumor in the upper figure shows lower NAA in tumor regions and higher relative Cho, whereas the stable tumor in the lower figure demonstrates higher NAA with fewer metabolic abnormalities. NAA is on the right, and tCho and total Cr are in peaks just left of the center as labeled. Spectra data were processed after phasing/frequency corrections and coil combination (with baseline, no quantification). The metabolite range is from ∼1.8 to 4.2 ppm. This material was obtained with permission and in collaboration with Yan Li and Peder Larson in the Department of Radiology and Biomedical Imaging at the University of California, San Francisco. tCr indicates total Cr.
Although there is growing literature on the safety of surgical implants at higher field strengths,160⇓⇓-163 there are not yet any definitive guidelines for imaging patients with nonmagnetic standard titanium plates placed during HGG resections, which has limited the clinical integration of 7T MRS thus far.164 It is important to perform additional testing and develop explicit guidance on whether surgical implants are safe at higher fields.165 The increased use of 7T MRS to scan patients in this category would allow longitudinal evaluations of patients with HGG at different phases of treatment, including postoperative follow-ups during radiation therapy, which have largely been confined to lower field strengths but ought to be expanded more widely to higher field strengths.
Ascertaining absolute metabolite concentrations would make metabolic information more accurate.166 Measuring T1 and T2 values is one of the only ways to acquire these concentrations.28 Ratios are derived using Cr peaks, which often vary depending on tissue type, making precise measurements difficult. Unsuppressed water-reference scans provide another way to more accurately quantify concentrations that is superior to ratios because they account for the relaxation times of both metabolites and of water voxels.55,167⇓⇓-170 Additional standardized markers of tumor aggressiveness would also be useful.171 Intracellular pH, energy metabolism, and total Cho may indicate more aggressive behavior in gliomas but must be verified.28 Other metabolites may also warrant further investigation. These include GSH, challenging even for 7T MRS to quantify. There have been a few reports of increased GSH compared with total Cr in HGGs, but there are less data supporting this trend.31 Given that this metabolite protects cells from free radicals, GSH might be a meaningful therapeutic target.172,173 Increasing the affinity of 7T MRS for GSH might, therefore, be worthwhile. Most important, GSH and other metabolites including GABA can be visualized at 3T with spectral editing techniques such as MEGA-PRESS.174 These editing techniques can provide better results at 7T because the increased spectral dispersion allows greatly reduced contamination from coedited macromolecules due to the bandwidth of the editing pulses. High-field acquisitions not only offer higher SNR but also address the problem of overlapping metabolites, better defining low-concentration metabolites such as GSH.31,175 Future studies should explore integrating MEGA-PRESS into 7T MRS to better visualize GSH.
Another metabolite for which further study may be of benefit is Lac. Although its significance in HGGs and other brain tumors is still not entirely clear, Lac reflects anaerobic glycolysis, which may be a sign of neoplastic growth.176,177 Increased glycolysis and perfusion in tumor tissue lead to elevated Lac, with higher levels corresponding with increased aggressiveness.178,179 Imaging Lac in HGGs via techniques such as MRS is crucial because this metabolite may be a biomarker of more aggressive tumors, with increased Lac correlating particularly strongly with grade IV gliomas.180 Although there have been several studies using lower-field-strength MRS to detect increased Lac, there is still very little 7T MRS literature on Lac in human subjects.27,34,181 Further investigation is warranted to determine whether 7T provides better visualization of Lac, which could serve as a lens into malignant activity, helping clinicians determine glioma grades, identify the best approaches to treatment, and monitor tumors across time.
Larger signal loss from T2 relaxation resulting in diminished SNR has also been registered at 7T, which necessitates approaches such as stimulated echo acquisition mode sequences with ultrashort TEs and longer TRs, which are optimal for tissues with unknown metabolic relaxation rates.33,182 Although 7T MRS can resolve many overlapping resonances and has even been shown to separate Glu from Gln, further work is necessary to differentiate metabolites such as these 2 while maintaining high spatial resolution and SNR.31 Moreover, 7T MRSI often fails to localize lipids in necrotic regions.31 Future studies will correct these deficiencies and capitalize on the power of 7T to distinguish abnormal metabolites.
CONCLUSIONS
We summarized the literature on 7T MRS and affirmed its usefulness in measuring metabolites. 7T MRS can achieve superior SNR and spectral separation and modestly improved spatial resolution and can also resolve overlapping resonances and isolate specific metabolites such as Glu, Gln, and Gly, which may be valuable therapeutic targets, improving patient outcomes. 7T MRS can also result in reduced CRLB values and improved quantification of small signals compared with 3T MRS. The increased spatial resolution, SNR, and spectral separation of 7T MRS may also enhance the detection of infiltrated tissue and treatment effect. We then underscored the hallmarks of effective 7T spectroscopy sequences that maximize SNR and spatial resolution and minimize B0 and B1 inhomogeneity, RF, and acquisition time.
We conclude with some of the failings of 7T MRS. These include safety concerns, the lack of data on other promising metabolites such as GSH and Lac, and the absence of absolute metabolite concentrations and established indices of aggressiveness. These flaws will be addressed as knowledge of glioma pathology and imaging technology continues to evolve.
Acknowledgments
We thank Drs Peder Larson, Yan Li, Sanjeev Chawla, and Suyash Mohan for helpful correspondence and images for figures.
Footnotes
Disclosure forms provided by the authors are available with the full text and PDF of this article at www.ajnr.org.
Priti Balchandani and Constantinos G. Hadjipanayis are co-senior authors.
This article was funded by a National Institutes of Health grant, R01CA202911.
Indicates open access to non-subscribers at www.ajnr.org
References
- 1.↵
- 2.↵
- 3.↵
- 4.↵
- 5.↵
- 6.↵
- 7.↵
- 8.↵
- 9.↵
- 10.↵
- 11.↵
- 12.↵
- 13.↵
- 14.↵
- 15.↵
- 16.↵
- 17.↵
- 18.↵
- 19.↵
- 20.↵
- 21.↵
- 22.↵
- 23.↵
- 24.↵
- 25.↵
- 26.↵
- 27.↵
- 28.↵
- 29.↵
- 30.↵
- 31.↵
- 32.↵
- 33.↵
- 34.↵
- 35.↵
- 36.↵
- 37.↵
- 38.↵
- 39.↵
- 40.↵
- 41.↵
- 42.↵
- 43.↵
- 44.↵
- 45.↵
- 46.↵
- 47.↵
- 48.↵
- 49.↵
- 50.↵
- 51.↵
- 52.↵
- 53.↵
- 54.↵
- 55.↵
- 56.↵
- 57.↵
- 58.↵
- 59.↵
- 60.↵
- 61.↵
- 62.↵
- 63.↵
- 64.↵
- 65.↵
- 66.↵
- 67.↵
- 68.↵
- 69.↵
- 70.↵
- 71.↵
- 72.↵
- 73.↵
- 74.↵
- 75.↵
- 76.↵
- 77.↵
- 78.↵
- 79.↵
- 80.↵
- 81.↵
- 82.↵
- 83.↵
- 84.↵
- 85.↵
- 86.↵
- 87.↵
- 88.↵
- 89.↵
- 90.↵
- 91.↵
- 92.↵
- 93.↵
- 94.↵
- 95.↵
- 96.↵
- 97.↵
- 98.↵
- 99.↵
- 100.↵
- 101.↵
- 102.↵
- 103.↵
- 104.↵
- 105.↵
- 106.↵
- 107.↵
- 108.↵
- 109.↵
- 110.↵
- 111.↵
- 112.↵
- 113.↵
- 114.↵
- 115.↵
- 116.↵
- 117.↵
- 118.↵
- 119.↵
- 120.↵
- 121.↵
- 122.↵
- 123.↵
- 124.↵
- 125.↵
- 126.↵
- 127.↵
- 128.↵
- 129.↵
- 130.↵
- 131.↵
- 132.↵
- 133.↵
- 134.↵
- 135.↵
- 136.↵
- 137.↵
- 138.↵
- 139.↵
- 140.↵
- 141.↵
- 142.↵
- 143.↵
- 144.↵
- 145.↵
- 146.↵
- 147.↵
- 148.↵
- 149.↵
- 150.↵
- 151.↵
- 152.↵
- 153.↵
- 154.↵
- 155.↵
- 156.↵
- 157.↵
- 158.↵
- 159.↵
- 160.↵
- 161.↵
- 162.↵
- 163.↵
- 164.↵
- 165.↵
- 166.↵
- 167.↵
- 168.↵
- 169.↵
- 170.↵
- 171.↵
- 172.↵
- 173.↵
- 174.↵
- 175.↵
- 176.↵
- 177.↵
- 178.↵
- 179.↵
- 180.↵
- 181.↵
- 182.↵
- Received November 3, 2021.
- Accepted after revision February 11, 2022.
- © 2022 by American Journal of Neuroradiology