Abstract
BACKGROUND AND PURPOSE: Different MR pulse sequences have been proposed for measuring multiple sclerosis (MS)-related abnormalities. The reproducibility of measured brain MS lesion volumes was compared for MR images performed using different scanners and different pulse sequences.
METHODS: Nine patients with relapsing-remitting MS were each imaged on two scanners and, on each occasion, dual-echo conventional spin-echo, dual-echo rapid-acquisition relaxation-enhanced (RARE), and fast fluid-attenuated inversion recovery (fast-FLAIR) images were obtained. The lesion volume present on each image was evaluated three times by a single observer in random order, using a local thresholding technique.
RESULTS: The mean lesion volumes present on fast-FLAIR images were significantly higher than those measured on dual-echo conventional spin-echo and RARE images. The mean intraobserver coefficients of variation for the different sequences and scanners ranged from 3.0% to 4.2% (no statistically significant difference). For each of the sequences, the use of different scanners introduced a variability that was higher than the intraobserver variability: the interscanner coefficient of variation was 7.4% for conventional spin-echo, 9.5% for RARE, and 18.5% for fast-FLAIR images.
CONCLUSION: Our study confirms that the use of different scanners significantly influences lesion loads measured from MR images of patients with MS and establishes that newer sequences are more susceptible to measurement variability. It also indicates that, if newer sequences are to be used in clinical trials, careful standardization is needed.
Changes of lesion load on yearly dual-echo brain MR images are used as a secondary end-point to monitor the effects of treatment on multiple sclerosis (MS) evolution in large-scale phase III clinical trials (1). Given the expected lesion load change in untreated patients with MS, which is on average approximately 5% to 10% per year (2), the procedure used for the lesion load must be highly reproducible. Although several factors influence lesion load measurements in MS (3–5), only the variability introduced by the human operator who performs the measurements has been studied in detail, by repeatedly measuring the same images from a single scanner (5). Since recent studies have reported that scanner performance (6) and accuracy in repositioning (7–10) are among the major contributors to measurement variability, the validation of lesion load assessment should include evaluating scan-rescan variability on the same and, when possible, on different scanners. During multicenter MS clinical trials with follow-up periods of 2 to 3 years, major upgrades of some of the MR scanners used are likely to occur.
Previous studies (11–15) have reported that rapid acquisition relaxation-enhanced (RARE) sequences detect virtually the same MS lesion numbers and loads as do conventional spin-echo (CSE) sequences, with comparable intra- and interobserver variability. The advantage of RARE is that dual-echo images are acquired in substantially less time than when using CSE. Findings of some recent studies (12, 13, 15–17) have suggested that fast fluid-attenuated inversion recovery (fast-FLAIR) sequences detect more MS lesions and result in larger lesion loads with lower intra- and interobserver variability than when using CSE. Nevertheless, a greater potential for variability of sequence design was found for the more complex, faster sequences, which could influence variability when different scanners are used. This variability, which has been determined to be significant for CSE images (6), has been evaluated in the present study.
Methods
Patients
Nine outpatients (five men and four women) with clinically definite MS (18) were included in the study. Five patients were recruited in Rome and four in Milan. All had relapsing-remitting disease courses (19). Their mean age was 29 years (SD, 5.5 years), median duration of the disease was 6 years (range, 3 to 10 years), and median Expanded Disability Status Scale (EDSS) (20) score was 1.5 (range, 1.0 to 2.5). Informed consent was obtained from all patients before inclusion in the study.
MR Protocol
During two sessions (one held in Milan and one in Rome) separated by an interval of 18 to 24 hours, patients were imaged using two MR units operating at 1.5 T (Siemens Vision and Philips Gyroscan NT). The two MR units will be referred to as scanner A and B to present the results anonymously. All images consisted of 44 contiguous, interleaved, 3-mm-thick axial sections with a rectangular field-of-view of 250 × 188 mm (anteroposterior × left-right). One acquisition was used for all sequences (apart from sequence f below, for which two acquisitions were used). Dual-echo CSE, dual-echo RARE, and fast-FLAIR images were obtained for all patients. The patients were accurately repositioned according to guidelines established by a European Community Committee for MS (21). The following acquisition parameters were used:
sequence a, CSE/scanner A: 2200/20,80/1 (TR/TE/excitations); raw-data matrix = 256 × 192; in-plane resolution = 0.98 × 0.98 mm; acquisition time = 14 minutes 12 seconds;
sequence b, CSE/scanner B: 2700/30,90/1; raw-data matrix = 256 × 145; in-plane resolution = 0.98 × 1.30 mm; acquisition time = 13 minutes 3 seconds;
sequence c, RARE/scanner A: 3800/22,90/1; echo train length = 5; raw-data matrix = 256 × 190; in-plane resolution = 0.98 × 0.98 mm; acquisition time = 4 minutes 56 seconds;
sequence d, RARE/scanner B: 3800/25,100/1; echo train length = 6; raw-data matrix = 256 × 192; in-plane resolution = 0.98 × 0.98 mm; acquisition time = 3 minutes 55 seconds;
sequence e, fast-FLAIR/scanner A: 9999/105/1; TI, 2200; echo train length = 7; raw-data matrix = 256 × 182; in-plane resolution = 0.98 × 1.03 mm; acquisition time = 9 minutes 38 seconds;
sequence f, fast-FLAIR/scanner B: 6500/150/2; TI, 2000; echo train length = 19; raw-data matrix = 256 × 190; in-plane resolution = 0.98 × 0.99 mm; acquisition time = 6 minutes 56 seconds.
CSE and RARE acquisition parameters were optimized before study initiation to obtain similar contrast between normal tissues and MS lesions at both centers (Fig 1). At each center, the acquisition parameters used for the fast-FLAIR sequences were those considered optimal by the local investigators on the basis of their personal experience and were routinely used for clinical examinations. The two fast-FLAIR sequences resulted in similar contrast between normal tissue and MS lesions at the two centers (Fig 2). To check whether the use of these different fast-FLAIR sequences influenced the measured lesion loads, the two fast-FLAIR sequences were performed in another five patients, with the same clinical characteristics of those scanned in the main study. This was done only on scanner B, since scanner A was not capable of performing sequence f without modifications beyond the capabilities of most clinical sites. Both MR scanners were on a regular course of maintenance when the study was performed.
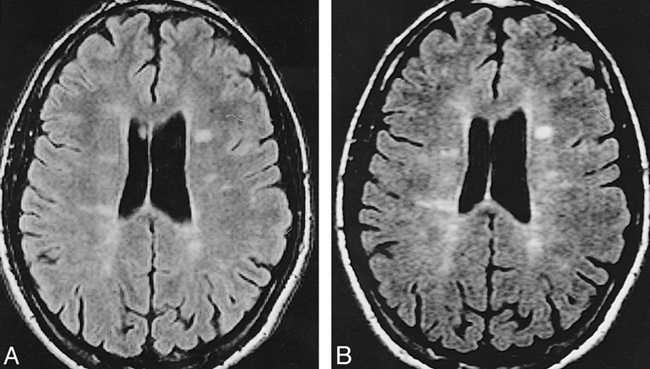
Axial RARE moderately T2-weighted images obtained from scanner A (A) (3800/22/1) and scanner B (B) (3800/25/1). Several hyperintense lesions are visible in both the cerebral hemispheres
Axial FLAIR images obtained from scanner A (A) (9999/105/1; TI, 2200) and scanner B (B) (6500/150/2; TI, 2000) at the same anatomic level as in figure 1
Quantification of MR Abnormalities
To evaluate the intraobserver and interscanner coefficients of variation (COVs), all of the MR abnormalities present on each image were measured three times by a single observer in random order without knowing the patient to whom the images belonged or the MR unit used, using a quantitative semiautomated technique based on local thresholding (22) and following published guidelines for MR lesion load quantification in MS (22). An interval of 1 month separated each of the measurement sessions. Intraobserver variability was defined as the variability between estimates of the lesion volume determined by a single observer who repeatedly evaluated the same images obtained for the same patients using the same MR scanner and the same sequence. Interscanner variability was defined as the variability between mean estimates of lesion volume determined by the same single observer when images of one patient obtained using the two different MR scanners and the same sequence type were evaluated. Thus, the interscanner variability includes not only the intraobserver variation but also the repositioning variability and the variation in observed lesion volume caused by different scanner hardware and sequence implementations. For the dual-echo images, the abnormalities were measured on the first-echo, moderately T2-weighted images (which had good definition of the MS lesions with some suppression of the CSF signal), using the second-echo, heavily T2-weighted images as a reference. Using the same segmentation technique, a single observer, without knowing the patient to whom the images belonged or the sequence characteristics and in random order, measured the lesion volumes from the five patients from whom the two fast-FLAIR images were obtained using scanner B.
Statistical Analysis
The components of variance (intraobserver and interscanner) were estimated using a mixed analysis of variance model (BMDP software, version 8 [BMDP Statistical Software, Cork, Ireland]). The intraobserver variances estimated in the different experimental situations were transformed into a variate with an approximate gaussian distribution by a logarithmic transformation and then compared using a classic linear model according to the method of Bossi and Milani (23). This procedure is the same used for the analysis of the effects on means, once the distribution of variances has been taken into account. The standard errors for the components of variances were estimated using the bootstrap resampling technique (24) implemented on the SPLUS (Statistical Sciences, Inc, Seattle, WA) random number generator. The degrees of freedom for the components of variance were estimated by the Satterthwaite method (25). Interscanner variances were compared using the Bartlett test for homogeneity.
Results
The overall means and standard errors of the lesion volumes obtained for the entire sample and those for each technique and scanner separately are presented in Table 1. The overall intraobserver COVs and the COV for each technique and each scanner are presented in Table 2. The mean intraobserver COV for the different sequences and scanners ranged from 2.6% (for RARE images obtained on scanner A) to 4.2% (for CSE images obtained on scanner B). Nevertheless, the intraobserver COV obtained using different scanners (P = .18) and sequences (P = .09) did not differ significantly.
Mean (standard error) lesion volumes (mL) obtained for different scanners and sequences
Mean (standard error) coefficients of variation (%) obtained for different scanners and sequences
The use of different scanners introduced a variability that was significantly higher than the intraobserver variability (P < .0001). However, this effect was not systematic (lesion volumes were higher on CSE and fast-FLAIR images obtained on scanner B, whereas the reverse was true for RARE images [Table 1]). The interscanner COVs were 7.5% (standard error = 1.5%) for CSE, 9.5% (standard error = 2.1%) for RARE, and 18.5% (standard error = 2.7%) for fast-FLAIR sequences (χ2 = 22.9, P < .01).
The lesion volumes measured on the images obtained using the two fast-FLAIR sequences on scanner B are presented in Table 3. In four of the five patients, lesion volumes were higher with the sequence routinely used in scanner B.
Lesion volumes (mL) obtained from five patients with MS undergoing imaging with the two fast-FLAIR sequences on scanner B
Discussion
Our results confirm that brain lesion volumes and measurement reproducibility in MS are markedly influenced by the use of different MR scanners. This influence is much greater than that caused by operator variability, and becomes more important for RARE and fast-FLAIR than for CSE sequences.
We detected variable lesion loads in the same patients when different pulse sequences were used, with the highest volumes measured on fast-FLAIR images from both scanners. This finding is in agreement with previous studies that compared lesion volumes measured on CSE, RARE, and fast-FLAIR images (13, 15–17) with 5-mm-thick sections, suggesting that MR acquisition procedures are available that enable a fuller assessment of the overall MS disease burden (26–28). In a previous study (6), we showed that, for CSE images, the lesion volume detected also increases with increasing field strength. Since the two scanners used in the present study both operate at 1.5 T, it is likely that even greater differences in lesion volume would be seen from the range of scanners typically used in clinical trials.
The intraobserver variability in measuring lesion load was similar to, if not better than, that seen in previous studies using segmentation techniques based on local thresholding (5). It was slightly better for fast-FLAIR images than for CSE and RARE images, probably because of the higher lesion conspicuity on fast-FLAIR images, which improves the performance of local thresholding segmentation techniques. The need for manual editing of poorly delineated lesions is thus reduced, leading to reduced measurement variability. The situation changes dramatically when evaluating interscanner variability. Interscanner variability depends on the variability of scanner performance and sequence implementation and on the reliability of patient repositioning. Since, in the present study, the three sequences were obtained in a single session on each scanner, we can assume that the variability in repositioning would similarly influence all three sequences. Thus, we can be sure that the higher interscanner variability of the fast-FLAIR sequence is attributable to the different acquisition parameters used. Specifically, the longer TE and echo train length used for acquiring fast-FLAIR images on scanner B resulted in a better lesion conspicuity, particularly for smaller lesions, thus influencing both measured lesion volumes and measurement reproducibility. While the TR for scanner B was considerably shorter than for scanner A, this was combined with a shortened TI to maintain good CSF nulling without compromising white matter/lesion contrast (29). It is likely that the situation would be improved by matching the fast-FLAIR sequences more closely. However, it is possible that scanner-specific constraints limit the degree to which sequences can be matched, a problem that would be encountered when planning a multicenter longitudinal study. For example, a limited choice of echo train lengths and TEs was found for scanner A used in this study, which prevents the use of a fast-FLAIR sequence that has close to optimum parameters (29). While the TR for the CSE sequences differed considerably between the two sites, the CSE lesion load measurements had the greatest interscanner consistency. Thus, it would seem that the lesion loads visible on CSE images are more tolerant of slight variations in contrast, and that the simplicity of this sequence does not allow great implementational differences between scanners of different manufacturers to arise.
Conclusion
The findings of this study confirm that the use of different scanners influences significantly the lesion load measurements from MR images of patients with MS, and that this influence is higher for more complex sequences. Without careful standardization, sequences such as fast-FLAIR might result in an unacceptable intersite variability. In future clinical trials, if more complex imaging sequences are to be used, the standardization and stability of sequence implementation will become a more important issue.
Footnotes
↵1 Supported by grants from the Istituto Superiore di Sanità (ISS, no. 96/J/T49) and by Associazione Italiana Sclerosi Multipla (AISM). M.P.S. was supported by a grant from TEVA Italy.
↵2 Address reprint requests to Massimo Filippi, MD, Neuroimaging Research Unit, Department of Neuroscience, Scientific Institute Ospedale San Raffaele, Via Olgettina, 60, 20132 Milan, Italy.
References
- Received April 21, 1998.
- Copyright © American Society of Neuroradiology