Abstract
Summary: We describe a method of converting postprocessed functional MR images to the Digital Imaging and Communications in Medicine (DICOM) standard and sending these DICOM images directly into any picture archiving and communication system (PACS) or stand-alone DICOM database. This method provides system-wide access and archiving of previously research-only applications, it permits the clinical review of postprocessed data on DICOM-compliant workstations, and it can be used to move functional MR data onto intraoperative neuronavigational workstations for surgical guidance. The procedure can be used with any MR postprocessed dataset, and it can be extended to other imaging modalities.
The conventional paradigm for dealing with functional MR imaging (fMRI) data, or research MR applications in general, involves sending imaging data acquired from the MR system to an off-line workstation on which all postprocessing is performed. The postprocessing of fMRI data frequently includes reconstructing raw data files into images, extensive statistical modeling, and integrating these with structural MR images in the form of color-coded overlays. In most cases, this workstation becomes the final resting place for the data. Although this method may be acceptable for basic research applications, it can become problematic in the setting of clinical research in which neuroradiologists, neurologists, and neurosurgeons have a keen interest in being able to review the data. Typically these workstations have a network connection to the magnets but no additional means of integration with existing installed picture archiving and communications systems (PACS), with image review stations, or with hard-copy film printers. MR manufacturers have addressed these issues in a limited way by providing some rudimentary fMRI postprocessing capabilities on commercial vendor-provided workstations and software. However, many investigators use specialized software packages or software developed in-house when they perform sophisticated postprocessing of datasets that cannot be integrated into the proprietary vendor-provided systems. Although the development of methods of integration is possible with proprietary vendor systems, this approach is tedious and depends on some degree of cooperation from the vendor in providing the source code or software support personnel. Furthermore, software revisions of these proprietary systems made by the vendor can rapidly make locally developed software obsolete.
Many institutions have already installed PACS for department-wide review and archiving of digital images. All MR manufacturers currently provide image review workstations that are compliant with the Digital Imaging and Communications in Medicine (DICOM) standard (1, 2). Ideally, postprocessed fMRI data would be available for review on these systems. In this article, we describe a robust method for integrating off-line fMRI data with PACS. This permits the system-wide availability of these data for review on the same workstations that are used in the clinical interpretation of conventionally acquired data. Furthermore this approach provides a much-needed bridge between the research and clinical domains in moving proof-in-principle research applications into the clinical realm. Our method was designed to be easily implemented on existing workstations, and it does not require the configuration of a DICOM database on the off-line workstation.
Technique
Background
The typical life cycle of fMRI data involves three phases: acquisition, postprocessing, and display. The acquisition phase occurs on the MR system and usually involves a raw data file. The raw data are then transferred to an off-line workstation for the remaining steps. The postprocessing can include the reconstruction of the raw data into images followed by the conversion of the images into some site-specific or software-specific format. At our institution, we convert the images to the ANALYZE format (Mayo Clinic, Rochester, MN), but we retain the raw data header information for a variety of automated functions. These ANALYZE images are then entered into an fMRI processing package that generates statistical maps as its output. These statistical maps are thresholded for significance, and surviving voxels are overlaid on the structural images with color coding to indicate areas of activation. These fused images are then displayed or rendered on the off-line workstation by using a variety of software programs. This is typically the endpoint of the fMRI dataset life cycle. For radiologists or clinicians to view these data, they must either physically come to the off-line workstation, or they must print a paper copy of the images on a color laser printer. Neither format is appropriate for standard clinical presentation, which involves the use of either a viewbox or a high-performance dedicated image-review station.
System Schematic
Our system operates entirely from an Ultrasparc platform running the Solaris operating system (SUN Microsystems, Palo Alto, CA). The fused fMRI datasets are written to ANALYZE format in IDL (Interactive Data Language Research Systems, Boulder, CO). We then convert these images to the DICOM format. The conversion occurs in two steps: header conversion and image overwrite. The header portion of a DICOM template image file is modified on the basis of the information in the fMRI acquisition raw data header and the header information in the anatomic images used for the overlay. The user is given the optional opportunity to modify some of the information fields in the DICOM header. The modifications are performed by using the freely distributed program dcm_modify_elements (available at www.erl.wustl.edu/DICOM/ctn.html). The image portion of the template is then modified using IDL. The modified DICOM images are then sent directly into a DICOM-compliant database (which includes any stand-alone DICOM viewing station, as well as any DICOM-compliant PACS database) by using another freely distributed program (send_image). The entire process is completely automated, with file conversion and data transfers occurring in the background.
DICOM Header Structure
Unlike most image formats, DICOM headers can be of variable size. This feature has been the main obstacle preventing the earlier implementation of a system as described here. Fields within the header are indexed by two numbers (in hexadecimal notation): a group and an element. An in-depth description of the DICOM header structure is beyond the scope of this discussion (1); however, several critical fields are used to uniquely identify any particular image as part of a series and examination prescription. These fields include the series instance unique identifier (UID), the study instance UID, and the service object pair (SOP) instance UID. These numbers consist of a root (eg, 1.2.840) that indicates the image to be DICOM, followed by a series of numbers indicating the manufacturer of the MR system, the examination, the series, and the image number. The numbers directly after the DICOM root, the DICOM object identifiers, are registered in the United States by the American National Standards Institute (ANSI). We have obtained our own DICOM object identifier for use with our DICOM conversion routines. In our implementation, we generate new UIDs by using our unique DICOM object identifier, followed by a series of numbers that index the accession number, series number, and image number. Modification of these UID fields is essential to prevent overwriting of the original images in the database. By using our own unique DICOM object identifier, we ensure that the native images in the PACS database will not be overwritten, and the source of these images is clear. Some of the fields in the header that we modify are listed in the Table.
Fields Modified in the DICOM Header
Central Test Node Software
The Mallinckrodt Institute of Radiology DICOM Central Test Node (CTN) software is a DICOM implementation that was designed to be used at the annual meetings of the Radiological Society of North America (RSNA) to foster cooperative demonstrations by the medical imaging vendors. This software is freely available for downloading at www.erl.wustl.edu/DICOM/ctn.html. For our implementation, we rely on two routines from the CTN package, dcm_modify_elements and send_image, which were compiled for the Solaris operating system. The first program, dcm_modify_elements, modifies the header portion of a DICOM image on the basis of the values read in from a text file. The text file lists the group, the element, and the updated value for the fields to be modified. The second program establishes the appropriate protocols for sending a DICOM image into a DICOM database.
Convert2Dicom
We have written a program within IDL, convert2dicom, that takes an ANALYZE image or a reconstructed image from a General Electric MR unit (GE Medical Systems, Milwaukee, WI) and converts it to a DICOM image file. The program reads the header information from the ANALYZE image or from the GE image and generates a text file that contains the group, element, and value information for the fields listed in the Table. When a GE header is provided to the program, all the DICOM fields listed in the Table are appropriately filled, including those reflecting patient-position coordinates within the magnet. The program opens a graphical user interface that shows some of the textual header information to be modified, giving the user an opportunity to make modifications to patient history and series descriptor fields before generating the text file. A text file is generated for each image on the basis of the fields listed in the Table, along with the appropriate field value. For example, the entries for patient name and patient identifier (ID), respectively, would look like the following: 0010 0010 “Doe, John” and 0010 0020 “12345678.”
The program then calls the dcm_modify_elements program from the CTN package with a template DICOM image file and the newly generated text file as inputs. This process generates a new DICOM file consisting of the modified header (with the field sizes and offsets in the header appropriately adjusted) attached to the template image. The template we use is a 256 × 256 16-bit integer DICOM image file with its patient-sensitive information stripped. The header stripping was previously accomplished by using the dcm_modify_elements routine. To accommodate images of different dimensions (ie, other than 256 × 256 16-bit integer), the “PXL Pixel Data” field (group 7fe0, element 0010) in the header must be modified to indicate the size of the image. The dcm_modify_elements routine does not allow modification of this field. However, because this field is the last element of the header before the actual image data, this modification can be done directly on the header within IDL. The header offset into the DICOM file for the location of the pixel data field can be easily determined from the size of the stored image. After the header portion of the file is modified for image size and rewritten, convert2dicom appends the image data to the new DICOM file. We have successfully used this method to convert GE format image files, ANALYZE format image files, as well as image arrays stored in computer memory into DICOM image files.
Send_Image
The modified DICOM images can be sent directly into a PACS- or DICOM-compliant database by using the send_image routine. The send_image routine takes the image filename and the destination as command line inputs, as well as entries for host and destination applications entity titles. To send the data into a PACS database, the PACS server must have an entry for the host computer and a name identifying it as a recognized application. This information authenticates the source of the send_image call. Some stand-alone DICOM-compliant workstations accept incoming DICOM files without requiring authentication (ie, Advantage Windows workstations; GE Medical Systems).
fMRI Color Information
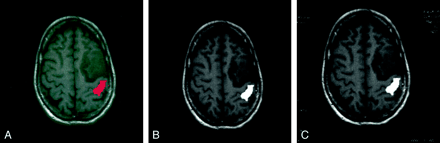
The fMRI data that we convert to the DICOM format consist of structural images with overlaid activations. On the off-line workstations, these images are displayed as gray-scale images with color overlays. In converting the images to DICOM, we have adopted the convention of scaling the signal intensities on the MR structural images within a range of 0–200 and assigning values of 200–255 for the activated voxels (ie, 210 for red, 220 for yellow). On gray-scale image-review stations, this method allows the activation to appear as white areas (Fig 1). Although the DICOM header structure can store information on color tables, as well as on individual color planes, at present, MR image review stations do not use any of the embedded color information.
fMRIs on an off-line workstation and on a PACS image review station.
A, fMRI from a unilateral hand motor task study displayed on an off-line workstation demonstrates activation of the primary motor cortex (red) posterior to a brain tumor.
B, Same image as in A, after DICOM conversion, displayed on an off-line workstation. Activated area is displayed in white by using a gray-scale color table. Otherwise, the images in A and B appear identical.
C, Same DICOM image as in B, after transfer to PACS image review station (Images are identical). In addition, the PACS image review workstation displays modified header fields, including the study date and image acquisition parameters (patient specific information was not filmed).
Placement of DICOM Images into Appropriate Patient Folders
DICOM studies sent to PACS are typically sorted by accession number (DICOM 0008, 0050). Our DICOM header fields are generated directly from the GE header information, including patient ID and accession number. These fields have been verified against the same images sent directly from the imaging unit into our PACS. We have adopted the convention of using the identifier fields associated with the anatomic image with which the functional information is merged. Thus, the modified DICOM image resides in the same study as the study in which the anatomic images were acquired. For our implementation, the series number field in the modified DICOM header and the DICOM UIDs are incremented beginning at 91 for axial overlays, 101 for sagittal overlays, and 111 for coronal overlays. For example, the third functional run in a study overlaid on axial anatomic images is assigned series number 93. The increment related to the overlay plane is not necessary; however, it ensures that an fMRI study overlaid on sagittal images does not receive the same study number as that of images from the same series that are overlaid on axial images. The critical feature is that native DICOM images in the PACS will not be overwritten. This protection is ensured by the use of our unique UIDs, even in the unlikely event that an MR examination has more than 90 series.
Validation of DICOM Header Information
The modified DICOM header fields listed in the Table are computed directly from the GE header information associated with the fMRI study and the anatomic overlay images. We have previously created IDL routines that read GE raw data headers (ie, Pfiles) as well as GE anatomic image file headers (ie, I.001 or E1234S2I4). Further discussion of these header-reading routines is beyond the scope of this report; however, they have been carefully validated for use in our fMRI applications.
Our fMRI analyses are performed in an automated fashion with regard to image orientation. This information is generated completely from the image headers, eliminating any user interaction in image orientation. To validate the accuracy of the modified fields in the DICOM headers, the values generated for our modified DICOM headers were verified against the same acquisitions sent from our GE machines into our PACS.
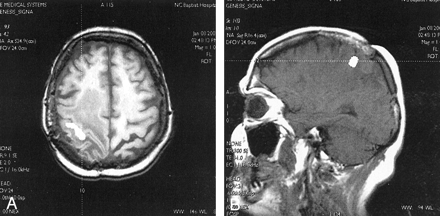
The image orientation fields were validated against images acquired in the sagittal, axial, and coronal planes. As further validation, we used the cross-referencing tools on our PACS with our modified DICOM files against images acquired in sagittal, axial, and coronal planes in the same study. Any errors in the image orientation fields would result in inaccurate cross-referencing to other planes (or the inability to cross-reference all together). The cross-referencing tools worked exactly the same on our modified sets and on the native datasets sent from our scanners into the PACS. Figure 2 demonstrates a cross-referencing example from a clinical case in which the functional data from a left-hand motor paradigm was overlaid on both axial T1-weighted images and sagittal T1-weighted images that were converted to DICOM and sent to our PACS.
fMRIs with axial and sagittal overlays displayed on a PACS image review station with cross-referencing.
A, fMRI overlaid on axial T1-weighted image demonstrates activation of the primary motor cortex (white area) with surrounding tumor and edema. A cross-reference tool (dotted line) on the PACS image review station was applied to the activated region in the right motor cortex.
B, Images from the same fMRI study overlaid on a sagittal T1-weighted MR image. A cross-reference tool from the axial image in A cuts through the motor cortex activation cluster on the right side of the sagittal volume (dotted line). Patient-specific information (ie, name, examination number, accession number) was not filmed.
Although we are confident of the integrity of the DICOM headers generated in this fashion from the GE images, we are less so in the case of ANALYZE images. Our routines can generate DICOM images from ANALYZE images off-line; however, the ANALYZE header fields lack critical information such as patient name, patient ID, accession number, image orientation, and acquisition parameters. When generating DICOM images from ANALYZE files, we give the user the opportunity to enter the patient’s name, ID, and history; the study description and date; and the series and accession numbers. With images generated in this fashion, however, the native image orientation and acquisition fields are not filled in properly.
Clinical Use
We have been using these tools for our clinical fMRI studies for more than a year. We routinely merge our postprocessed clinical fMRI images to the patient’s high-resolution T1-weighted images, convert them to DICOM format, and send the fused images into the PACS. Our neurosurgeons have been able to use these modified DICOM images for intraoperative guidance without difficulty. In addition, a neuroradiologist uses a PACS workstation to evaluate all the clinical fMRI studies that we send to our PACS; this step adds an additional layer of quality control to our clinical research operations.
Discussion
We have described a method of integrating fMRI data with a PACS database. This approach can be used with any postprocessed MR data, and it can be extended for use with other imaging modalities. Our method was designed to be easily implemented on existing workstations, and it does not require the configuration of a DICOM database on the off-line workstation. Unlike commercial DICOM PACS conversion solutions, the cost of implementing this technique would be negligible for most sites. The routines described here are written in IDL, or they are freely available on the Internet. Although a new single-user IDL license can cost approximately $1500, IDL is a common image processing language, and it is already available in many radiology departments and functional imaging laboratories. A similar method can be implemented by using C routines, which would obviate the purchase of an IDL license.
Currently, the major clinical application of fMRI is in presurgical mapping. The approach described here provides a robust solution for integrating the presurgical mapping fMRI data with intraoperative neuronavigational workstations for intraoperative guidance. These neuronavigational workstations provide the neurosurgeon with a probe-guided MR display of the brain in the operating room as the procedure is being performed. Current generation neuronavigational workstations accept DICOM images, obviating reverse engineering of the proprietary vendor header formats to integrate off-line fMRIs with these workstations, as described previously (3). When fMRIs are downloaded onto an intraoperative neuronavigational workstation, they necessarily lose color information (with any scheme) unless some proprietary solution is achieved with the specific workstation vendor.
To move MR research applications into the clinical realm, postprocessed images must be available on the same image review stations that radiologists and clinicians use to view conventional clinical imaging studies. This setup allows department-wide access to the images, permits hard-copy filming of the images, and allows a method of image archival for future review or comparison with previous studies. Furthermore, because all commercial MR units and PACS are DICOM compliant, this method is not at risk of becoming operationally challenged with any vendor software revisions.
Footnotes
Supported by the American Roentgen Ray Society Scholars Program 1999–2000.
- Received January 22, 2001.
- Accepted after revision April 12, 2002.
- Copyright © American Society of Neuroradiology