Abstract
BACKGROUND AND PURPOSE: There is significant interest in the development of novel noninvasive techniques for the diagnosis of Alzheimer disease (AD) and tracking its progression. Because MR imaging has detected alterations in sodium levels that correlate with cell death in stroke, we hypothesized that there would be alterations of sodium levels in the brains of patients with AD, related to AD cell death.
MATERIALS AND METHODS: A total of 10 volunteers (5 with mild AD and 5 healthy control subjects) were scanned with a 20-minute sodium (23Na) MR imaging protocol on a 3T clinical scanner.
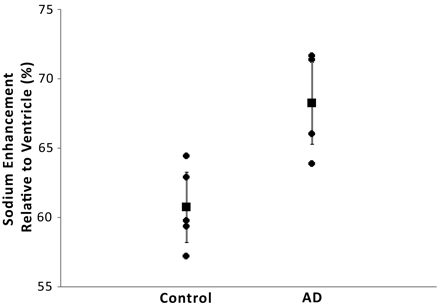
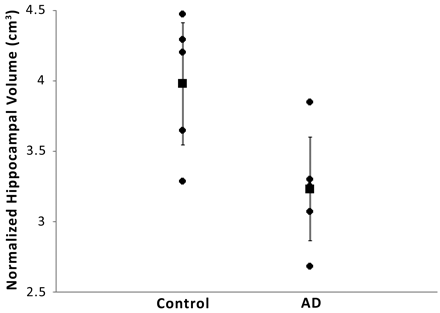
RESULTS: After normalizing the signal intensity from the medial temporal lobes corresponding to the hippocampus with the ventricular signal intensity, we were able to detect a 7.5% signal intensity increase in the brains of patients with AD (AD group, 68.25% ± 3.4% vs control group, 60.75% ± 2.9%; P < .01). This signal intensity enhancement inversely correlated with hippocampal volume (AD group, 3.22 ± 0.50 cm3 vs control group, 3.91 ± 0.45 cm3; r2 = 0.50).
CONCLUSIONS: This finding suggests that sodium imaging may be a clinically useful tool to detect the neuropathologic changes associated with AD.
Alzheimer disease (AD) is a devastating late-life dementia that produces progressive loss of memory and mental faculties in elderly people. More than 5 million people in the United States have AD, leading to a total health care cost of more than $50 billion USD annually.1 There is significant interest in biomarkers that can identify the earliest evidence of AD to allow for early initiation of treatment, quantitatively evaluating new interventions, and tracking a patient's response to treatment.2 To these ends, many approaches have been proposed, including the sampling of CSF for biochemical analysis of disease markers,3 positron emission tomography,4 and MR imaging.5 Because MR imaging is noninvasive and is routinely performed in the setting of neurologic disease, implementation of new MR imaging–based techniques for the diagnosis and prognosis of AD is especially advantageous.
There is an expanding collection of quantitative MR imaging–based techniques for the detection of AD. The most mature of these techniques is the regional volumetric analysis of brains from patients with AD compared with the brains of elderly control subjects. Previous studies have shown a reduction in hippocampal6 and cortical7 volume with related expansion of ventricular volume. More recent studies have tracked rates of whole-brain8 and hippocampal9 atrophy in AD and showed changes in hippocampal rate of atrophy during trials of an investigational AD drug.10 Still, numerous other techniques to measure changes in AD with MR imaging also show promise. As examples, MR spectroscopy has shown decreased levels of N-acetylaspartate11 in the AD brain that increase with treatment.12 Studies on mouse models have shown changes in T2,13,14 and T1ρ relaxation.15 However, human research with T2 relaxation has been conflicting16-18 and studies with changes in T1ρ are underway.19 Also, decreased cerebral blood flow20 and changes in magnetization transfer contrast21 have been demonstrated with MR imaging in AD. These techniques, either alone or in combination, warrant additional study for clinical use in tracking AD.
All of these MR imaging techniques rely on the proton nuclear spin resonance to make measurements. However, the spin resonances of other nuclei can be used. For example, reduced glutamate neurotransmission has been shown in patients with AD by 13C spectroscopy.22 Spectroscopy with 31P has shown alterations in the levels of membrane phosphate–containing compounds in AD.23 A compound labeled with fluorine was used to tag amyloid beta plaques, a histologic hallmark of AD, for 19F MR imaging studies in animal models.24
Another nucleus that has been used for in vivo MR imaging is the 100% naturally abundant sodium nucleus, 23Na. There are several demonstrated potential human disease applications for 23Na imaging. In cartilage, 23Na concentration has been correlated with proteoglycan content, an indicator of joint health.25,26 Cellular viability in myocardial ischemia has been predicted by looking at the changes in intracellular 23Na.27 In the brain, 23Na has been used to differentiate tumors from surrounding tissue28,29 as well as track cell death in cerebral infarction.30,31
In many of these applications, sodium MR imaging has been shown be sensitive to cell death and viability.32,33 Alzheimer disease is a disease where neural cell death has been well established. As a result, we tested whether changes would be seen in AD, with emphasis to the areas most affected by AD. In this study, we demonstrate for the first time with sodium MR imaging changes in sodium in patients with AD compared with age-matched control subjects.
Materials and Methods
Statistics were performed in Excel 2007 (Microsoft, Redmond, Wash) or SPSS version 16 (SPSS, Chicago, Ill). All P values are the results of 2-tailed independent-sample t tests assuming unequal variance. All r2 values are the square of the Pearson correlation from a bivariate correlation, unless otherwise noted.
Subjects
Subjects were selected from patients and cognitively normal subjects participating in the Alzheimer Disease Center (ADC) longitudinal clinical cohort. Clinical diagnosis (control and probable AD) and willingness to participate were the only selection criteria. Ten elderly volunteer participants underwent MR imaging: 5 mildly impaired patients with a clinical diagnosis of probable AD (Mini-Mental State Examination [MMSE] score: 18–28; mean, 24) and 5 age-matched healthy elderly control subjects (MMSE, 30 in all). Subjects were closely age and sex matched (control mean age, 74.4 years; SD, 5.4 years; range, 66–81 years; 4 men; AD mean age, 73.6 years; SD, 5.5 years; range, 64–77 years; 3 men). All received a standardized evaluation as specified by the National Alzheimer's Coordinating Center (NACC).34 This evaluation included the collection of medical information; performance of a standardized physical and neurologic examination; and cognitive testing with the NACC Uniform Data Set psychometric battery to determine impairments in memory, language, constructional praxis, and executive function, as well as standardized ratings of mood and behavior.35 The degree of global impairment was assessed with the Clinical Dementia Rating.36 Laboratory tests (eg, blood and urine studies, MR imaging, CSF tau, beta-amyloid levels, fluorodeoxyglucose–positron-emission tomography) were done at the discretion of the evaluating clinician. The final diagnosis for each subject was established at the weekly ADC diagnostic consensus conference after presentation of all relevant data by the evaluating clinician. All subjects provided informed consent, approved by our institutional review board, to participate in this study.
MR Imaging
Imaging was performed with a broadband-enabled 3T Trio clinical scanner (Siemens, Erlangen, Germany). A series of scout images were first obtained in a vendor-supplied 8-channel head array coil. These were used to position an isotropic T1-weighted inversion recovery–prepared 3D gradient–recalled echo (GRE) sequence with the following parameters: FOV, 245 × 245 mm; matrix, 192 × 192; section thickness, 1.28 mm (ST); TR, 1620 ms; inversion recovery time, 950 ms; flip angle (FA), 15°; 6/8 partial Fourier (PFT) in phase encode and section encode dimensions; and scan time, 3 minutes.
The patient was then placed into a custom-built 25-cm diameter, 20-cm length, quadrature birdcage head coil with a detached radiofrequency end cap37 tuned to the 23Na resonance of 32.6 Mhz. The coil was interfaced to the scanner with a custom transmit-receive switch. A low resolution, 12-second, 3-plane sodium fast GRE localizer was performed for section positioning. From this, the ventricles were clearly visible, and an outline of the head was seen to roughly position the section in the same way for each volunteer for the following coronal sodium head scan. A 0.146-cm3 voxel size, 20-minute coronal sodium-spoiled fast GRE scan was then performed with the following parameters: FOV, 245 × 245 mm; matrix, 64 × 64, ST, 10 mm; TR, 9.13 ms; TE, 2.96 ms (asymmetric readout); FA, 60°; 18 sections; nonselective excitation by a 1-ms hard pulse; 6/8 PFT (phase and section); elliptical scanning, 130 Hz/Px; and averages, 210.
A B1 map was generated with an 18-cm spherical 300-mmol/L NaCl phantom by running a series of 5 fast GRE scans each with a different applied transmitter voltage to generate FAs of 50, 70, 90, 110, and 130° when averaged over the entire volume. This was performed with the same parameters as the imaging in the previous paragraph but with TR 180 ms to allow for more complete T1 recovery, and interpolated in-plane to match the images in Fig 1A. Correction on the basis of the B1 map was not necessary because of the high homogeneity in the regions of interest.
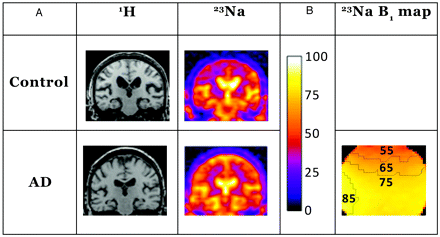
A, Representative coronal control (control C2) and AD (patient AD4) images are shown from the sections for region-of-interest analysis. The proton images demonstrate the anatomy of the brain in the corresponding sodium sections and were part of the dataset used for hippocampal volume determination. The sodium images are normalized to the ventricular signal intensity, scaled as shown with the color bar on the right, which is also used for B. The images were normalized to the brightest areas of intensity in the center of the image, seen clearly in these images as the lateral ventricles. The hippocampal volumes from the proton sets were coregistered to the sodium images and used to mask the hippocampuses as described in the methods. B, Shown is the corresponding section from a B1 map calculated from a spherical sodium phantom with contour lines added for each 10th percentile. This indicates excellent homogeneity over the areas of interest. The units are in B1 relative to the maximum in that section scaled to the same color bar. The maximum is found at the edges of the phantom near the struts of the birdcage.
Sodium Image Processing
The resulting k-space was reconstructed off-line with custom written software in IDL version 6.2 (ITT Software, Boulder, Colo). K-space filtering was performed with 3 (1 for each dimension) Gaussian functions, each with an alpha of 0.5, matrix multiplied together. This was selected among numerous filter choices because it provided the highest signal-to-noise ratio without noticeable loss of image detail from the sodium images. An automated algorithm took the average of the top 10% of pixel signal intensities in the area of the brain where lateral ventricles were expected from these unsmoothed images. The pixel intensities in each section were normalized to that ventricular signal intensity and checked for each brain. Figure 1 shows representative proton and sodium images from a control subject and a patient with AD as well as coil homogeneity.
Volume Computations
We removed skulls and surrounding tissues from the proton isotropic images by using the Brain Extraction Tool v1.3 (http://www.fmrib.ox.ac.uk/analysis/research/bet) and checked by using 3D visualization with MRIcro v1.4 (http://www.sph.sc.edu/comd/rorden/mricro.html). We computed total intracranial volume (TIV) by taking the total volume of all the remaining pixels, composed of brain and ventricle, from the bottom of the cerebellum to the top of the brain. We manually segmented the hippocampus using ITK-SNAP version 1.4 (http://www.itksnap.org/) according to the boundaries presented previously by Bartzokis et al38 by using the posterior boundary definition of Watson et al.39 To summarize, the most anterior section is where the alveus becomes visible, the most posterior section is the section where the crura of the fornices are seen in full profile, the lateral and inferior borders are the gray/white matter interface, the medial border is the CSF, and the subiculum and alveus are included in the measurement.
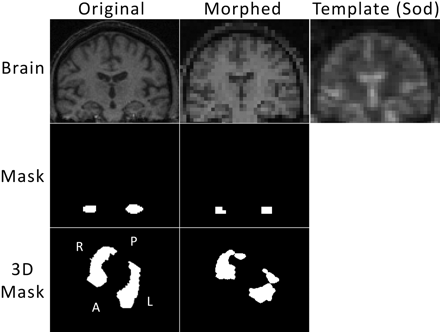
Because the sodium images offer less tissue contrast and lower resolution, the hippocampal regions obtained in the previous paragraph were used to define the areas of the hippocampus in the sodium images as follows. A rigid body co-registration was performed to match each volunteer's proton image set to their corresponding sodium image set with use of statistical parametric mapping version 5 (SPM5, http://www.fil.ion.ucl.ac.uk/spm/). These were resectioned to match the sodium voxel size. The hippocampal volume masks underwent the same co-registration and resection as the corresponding proton images. For 1 subject, this process is displayed in Fig 2. The average sodium enhancements with use of the co-registered, resectioned hippocampal masks on the sodium images are then reported.
For each subject, the proton images underwent 3D co-registration and resectioning to match the sodium position for that subject and sodium voxel size. The top 3 images show from left to right: 1 section of the original proton volume, the corresponding section of the co-registered and resectioned (Morphed) proton volume, and the corresponding section of the sodium volume for that subject. The hippocampal volumes were computed from manual hippocampal masking on a section-by-section basis. The middle left image shows the hippocampal mask for the section of brain shown above. The transformation matrix obtained from the co-registration and resection of the proton volume was applied to the hippocampal mask to yield the mask in the center image for that morphed section. This was performed on a 3D basis, and the bottom line shows that transformation in 3D. The entire hippocampal volume mask is shown as a 3D rendering of the surface from a tilted superior-inferior perspective. The morphed hippocampal volume mask (bottom center) was applied to the sodium volume, and those pixels were averaged to obtain sodium enhancement.
Results
The results of the analysis of the hippocampus-based regions of the medial temporal lobes of the control and AD cohorts are presented in the Table. Normalized overall enhancement was 60.75% ± 2.9% in the control group and 68.25% ± 3.4% in the AD group (P < .01). Hippocampal volumes, unnormalized and normalized to total intracranial volume (norm) were taken from T1-weighted brain images as described in the Methods section and are presented for comparison.
Cohort data summary
Sodium MR imaging correlates more strongly with AD diagnosis (Fig 3; r2 = 0.53; P = .006) than TIV normalized hippocampal volume (Fig 4; r2 = 0.42; P = .034) in this preliminary study. Not shown are the correlations for the ventricular normalized sodium then normalized to TIV and diagnosis (r2 = 0.24; P = .154) and unnormalized hippocampal volume and diagnosis (r2 = 0.39; P = .052).
Compared is sodium enhancement with diagnosis (r2 = 0.53; P = .006). Sodium enhancement is taken as the average sodium enhancement of the right and left medial temporal lobes relative to ventricle. The vertical error bars represent the 95% confidence interval.
Compared is diagnosis with mean hippocampal volume per subject normalized to total intracranial volume (r2 = 0.42; P = .034). The vertical error bars represent the 95% confidence interval.
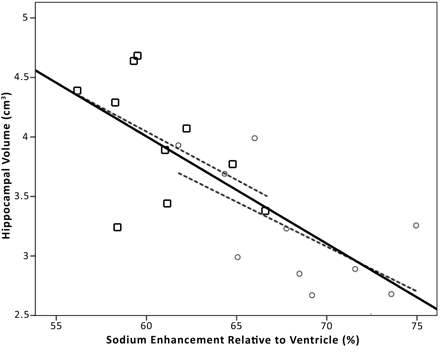
Figure 5 shows the hemisphere-to-hemisphere correlation between sodium enhancement and unnormalized hippocampal volume. A moderate correlation is observed overall (r2 = 0.50; P < .01). Correlations within the control (r2 = 0.24) and AD (r2 = 0.24) groups were also obtained. Sodium enhancement correlated to hippocampal volume normalized to TIV demonstrates weaker correlation overall (r2 = 0.27; P = .19), despite very similar pooled averages for unnormalized and normalized hippocampal volumes shown in the Table.
Plotted is unnormalized hippocampal volume against sodium enhancement for each brain hemisphere (20 hemispheres from 10 subjects; overall r2 = 0.50; P < .01). The square data points represent control hemispheres, and the circles represent AD hemispheres. The center solid line indicates the linear overall regression. The dashed lines indicate the linear regressions for the control (left: r2 = 0.24; P < .01) and AD (right: r2 = 0.40; P < .01) data points. Although a moderate correlation is observed for all groups, sodium enhancement still correlates with disease state even when controlling for hippocampal volumes.
To test whether the sodium signal intensity differences could be explained entirely by changes in hippocampal volume, we performed a partial correlation between disease status and sodium level, controlling for the unnormalized hippocampal volumes for each hippocampus. The correlation was still significant (P = .025), indicating that the sodium detection of AD cannot be entirely explained by differences in hippocampal volume.
Discussion
To our knowledge, sodium MR imaging has not been previously applied to the study of AD. To provide a comparison, we computed the hippocampal volume from these subjects from proton images to provide a more thoroughly tested AD biomarker. In this novel preliminary application of sodium MR imaging, the sodium MR imaging had a higher correlation with disease state than did hippocampal volume. In the literature and in our study, normalization to TIV improved correlation with disease state for hippocampal volume.40 However, normalization to TIV significantly weakened the sodium imaging values, suggesting that sodium MR imaging may provide different or complementary information about disease state. In a similar fashion, the measured sodium values correlated much more with unnormalized hippocampal volumes. This finding suggests that hippocampal processes directly contribute to the observed effect. Furthermore, controlling for those hippocampal volumes in the correlation of sodium enhancement to disease state does not invalidate the observed effect. This indicates that sodium MR imaging provides additional or different information beyond measurements of hippocampal volume.
Assumptions about the physiology and composition of brain tissue help to explain the observed results.41,42 In perfused brain tissue, the concentration of extracellular sodium, [Na]e, is in diffusional equilibrium with blood and CSF [Na] at a constant 140 mmol/L. The concentration of intracellular sodium, [Na]i, in the normal tissue is assumed to be approximately 12 mmol/L. The intracellular volume fraction (Vi) in gray matter is approximately 80%, whereas the extracellular volume fraction (Ve) is approximately 20%, and gray matter is the predominant tissue in our measured regions of interest.
In tissue sites where motional narrowing is achieved (ω0τc ≥ 1)τ, sodium exhibits monoexponential transverse relaxation. Biexponential transverse relaxation is observed in tissue sites where motional narrowing is not achieved (ω0τc ≥ 1); the slow component of relaxation, T2s, and fast component, T2f, comprise 40% and 60% of the total sodium signal intensity, respectively. The static quadripolar interaction is minor in the brain and can be effectively ignored.43 Averaged over the intracellular and extracellular compartments, T2s and T2f, have been estimated at 15 ms and 3 ms with a proportion of 40% and 60%, respectively, for normal gray matter.44,45 This can be split further into the intracellular components T2s,i and T2f,i and extracellular components T2s,e and T2f,e. The values of T2s,i and T2f,i have been approximated at their fastest from yeast cells as 7.5 and 0.35 ms,46 but also from rat liver as 13.0 and 1.3 ms.47 For extracellular fluid, T2s,e and T2f,e are thought to be significantly longer, perhaps on the order of 41.6 ms and 12.0 ms as estimated for blood serum42 or close to 18 ms for both components as reported for extracellular rat liver fluid.47 With the TE used in this study, most of the tissue sodium is visible except the component T2f,i, making the intracellular sodium approximately 30% visible (from the T2s,i component, accounting for a 3-ms decay of that component), accounting for approximately 13% of the overall sodium signal intensity (for 18-ms monoexponential extracellular relaxation). The relation of all of these factors to sodium signal intensity (Sna) is related by equation 1 where TE is the echo time of the sequence, A is a constant relating to a number of additional factors including the receptivity of sodium and the system and coil electronics, and it is assumed that the intracellular and extracellular pools each have 1 correlation time.  Physically and physiologically reasonable changes in the parameters of the equation would cause the observed increase in sodium signal intensity. Before cell death, increased sodium in tissue could be caused by direct leakage of sodium because of amyloid beta channels in membranes48 or by the impairment of the Na+/K+−ATPase.49 Given the large Vi of brain tissue, only a modest increase in intracellular sodium would be expected to produce this change in signal intensity, even with the partial visibility of intracellular sodium of this sequence. After neuronal death, the intracellular space shrinks and the extracellular space expands, and the larger sodium concentration in the extracellular space yields a larger sodium signal intensity. This hypothesis has been put forward to explain sodium changes in studies of cerebral infarction.31 In addition, with cell death are changes in tissue composition, potentially increasing the transverse relaxation times in AD-affected tissue. A combination of these effects is likely, and several experiments with multiple pulse sequences would need to be performed to separate and investigate all of these parameters.
Physically and physiologically reasonable changes in the parameters of the equation would cause the observed increase in sodium signal intensity. Before cell death, increased sodium in tissue could be caused by direct leakage of sodium because of amyloid beta channels in membranes48 or by the impairment of the Na+/K+−ATPase.49 Given the large Vi of brain tissue, only a modest increase in intracellular sodium would be expected to produce this change in signal intensity, even with the partial visibility of intracellular sodium of this sequence. After neuronal death, the intracellular space shrinks and the extracellular space expands, and the larger sodium concentration in the extracellular space yields a larger sodium signal intensity. This hypothesis has been put forward to explain sodium changes in studies of cerebral infarction.31 In addition, with cell death are changes in tissue composition, potentially increasing the transverse relaxation times in AD-affected tissue. A combination of these effects is likely, and several experiments with multiple pulse sequences would need to be performed to separate and investigate all of these parameters.
In our study, we chose a GRE sodium imaging sequence to obtain high signal-to-noise ratios (>20:1 from the regions of interest) and high-image quality with minimal image blurring and good anatomic localization. Rectilinear k-space sampling requires a TE that produces relaxation weighting and the loss of signal intensity from some fast-relaxing sodium nuclei. Therefore, the sodium enhancement measured in our study may be the result of increases in spin attenuation, changes in sodium relaxation times, or both. Center-out k-space trajectories reduce relaxation weighting and capture signal intensity from the fast-relaxing sodium spins, but they increase image blurring and are more technically difficult to implement. Therefore, we believe that the rectilinear k-space sampling may be the optimal approach to routine clinical sodium MR imaging. As examples of the usefulness of this approach, GRE enhancement of sodium measured with even longer TE has been used to track both cerebral50 and myocardial51 infarction in animal models. Furthermore, GRE measurements of sodium have been frequently encountered in the recent literature,52-56 and the sequence is easily run on any clinical scanner. It is more remarkable that even with this relatively long echo sequence, a potentially important diagnostic marker has been found.
Additional sodium studies, including on animal models of AD, can be performed to explore changes in every parameter in the equation to test the hypotheses put forward. Studies are currently in design to determine whether the major contribution to the signal intensity difference is because of changes in intracellular sodium concentrations, changes in the extracellular or intracellular volumes, or changes in the relaxation properties of sodium in the hippocampal areas. For example, intracellular sodium can be selected by inversion recovery56 or multiple-quantum filtering,57-59 and methods for total sodium measurements60 and relaxation mapping53 have been demonstrated in the literature.
Several challenges and limitations were presented in our current study. The first challenge was that any site that wishes to perform sodium imaging will need a sodium coil, scanner interface hardware, a broadband-enabled scanner capable of transmitting and receiving at sodium frequency, and a modified or custom pulse sequence for sodium. This may not be a long-term difficulty, as there is an increasing trend of availability of 3T MR imaging scanners with multinuclear capabilities. Sodium imaging fundamentally yields lower signal-to-noise ratios, and as such, higher-field clinical scanners of more than 3T are desired for their improved signal-to-noise ratio. An additional challenge was finding an appropriate standard for comparisons between subjects. Initial attempts were performed with external sodium-doped agarose reference phantoms; however, their position beside the head in close proximity to the struts of the birdcage coil caused unacceptable variations because of field inhomogeneity. To overcome this problem, we used the lateral ventricles as an internal control for sodium, as they were thought to contain a remarkably constant level of sodium very close to 140 mmol/L.61 In this case, it must be shown that the B1 profile of the coil is constant between the CSF and the compared tissue, or the inhomogeneity must be taken into account. To our knowledge, systematic differences in CSF sodium concentrations in neurodegeneration have not been reported.
Conclusions
Statistically significant changes in sodium signal intensity were observed between patients with mild AD and control subjects on a 3T clinical scanner. To our knowledge, this study is the first to demonstrate the feasibility of imaging AD with sodium MR. The 20-minute protocol shown here provides both good anatomic localization and signal-to-noise ratio. These sodium images can be signal intensity normalized for cohort studies, either with internal CSF/vitreous humor controls or external phantom references. A moderate inverse correlation was shown between sodium enhancement and measurements of hippocampal volume. These results demonstrate the potential of sodium MR imaging for early AD diagnosis in conjunction with anatomic proton MR imaging. We are currently exploring additional experiments with a variety of sodium pulse sequences to find the physiologic basis of these results. This would contribute to our understanding of factors responsible for the disease.
Acknowledgments
We thank Thomas J. Connick for his technical assistance with coil design and interface and Marianne Watson and Christa Nichols for their assistance with patient coordination.
Footnotes
Funding was provided by a National Center for Research Resources funded grant RR02305.
Previously presented in part at: Scientific Meeting and Exhibition of the International Society for Magnetic Resonance in Medicine, May 8, 2008, Toronto, Ontario, Canada; and International Conference on Alzheimer's Disease, July 26, 2008, Chicago, Ill.
References
- Received September 10, 2008.
- Accepted after revision December 6, 2008.
- Copyright © American Society of Neuroradiology