Abstract
BACKGROUND AND PURPOSE: Recent advances in machine learning have enabled image-based prediction of local tissue pathology in gliomas, but the clinical usefulness of these predictions is unknown. We aimed to evaluate the prognostic ability of imaging-based estimates of cellular density for patients with gliomas, with comparison to the gold standard reference of World Health Organization grading.
MATERIALS AND METHODS: Data from 1181 (207 grade II, 246 grade III, 728 grade IV) previously untreated patients with gliomas from a single institution were analyzed. A pretrained random forest model estimated voxelwise tumor cellularity using MR imaging data. Maximum cellular density was correlated with the World Health Organization grade and actual survival, correcting for covariates of age and performance status.
RESULTS: A maximum estimated cellular density of >7681 nuclei/mm2 was associated with a worse prognosis and a univariate hazard ratio of 4.21 (P < .001); the multivariate hazard ratio after adjusting for covariates of age and performance status was 2.91 (P < .001). The concordance index between maximum cellular density (adjusted for covariates) and survival was 0.734. The hazard ratio for a high World Health Organization grade (IV) was 7.57 univariate (P < .001) and 5.25 multivariate (P < .001). The concordance index for World Health Organization grading (adjusted for covariates) was 0.761. The maximum cellular density was an independent predictor of overall survival, and a Cox model using World Health Organization grade, maximum cellular density, age, and Karnofsky performance status had a higher concordance (C = 0.764; range 0.748–0.781) than the component predictors.
CONCLUSIONS: Image-based estimation of glioma cellularity is a promising biomarker for predicting survival, approaching the prognostic power of World Health Organization grading, with added values of early availability, low risk, and low cost.
ABBREVIATIONS:
- CD
- cellular density
- C-index
- concordance index
- KPS
- Karnofsky performance status
- max
- maximum
- ROC
- receiver operating characteristic
- WHO
- World Health Organization
The most powerful prognostic factor currently known for patients with gliomas is the tumor grade as described by the World Health Organization (WHO).1,2 The WHO grading system ranges from I to IV with a higher grade indicating increased malignancy and a worse prognosis. Historically, tissue histology has driven diagnosis and grading using characteristics like mitoses, microvascular proliferation, or necrosis.3 Recent updates emphasize molecular characteristics in WHO grading.1,2
WHO grading depends on having tissue specimens. Obtaining these specimens is difficult, expensive, and includes a risk for the patient. In current practice, diagnostic tissue samples are often obtained during the first surgical procedure, meaning that, in effect, a definitive tumor grade is obtained after some treatment decisions have already been made. When tissue is collected before bulk resection, it takes the form of small biopsy samples.4 All tissue-based approaches have some degree of risk with regard to sampling error and cannot capture the full range of heterogeneity present inside the tumor.
In contrast to tissue sampling, MR imaging is relatively inexpensive, safe, and easy to perform. Imaging does not have sampling error, and covers the whole brain, though not at the microscopic resolution of histology. Furthermore, imaging is available before the commencement of invasive therapies. Multiple imaging findings like contrast enhancement are strongly associated with a higher WHO grade5 and have proved very useful in the clinical management of these patients. However, most imaging findings are qualitative in nature and cannot yet replace WHO grading.
There is great clinical need for a noninvasive imaging tool that can accurately grade and stage patients with gliomas. One way is to estimate pathologic characteristics used in formulating tumor grade. Cellular density (CD) is increased in all gliomas and correlates with increasing WHO grades.2 CD is of additional clinical interest because the subtle infiltrative nature of diffuse gliomas, with increased CD blending into the healthy brain, makes these tumors difficult to treat. Several recent works have developed models capable of estimating heightened cellularity using MR imaging data.6-9 However, the actual prognostic value of these model estimates has not been directly validated.
In this study, we investigated image-based estimates of CD as a low-cost and low-risk predictor of overall survival for patients with gliomas. We correlated CD and the gold standard of histology-based WHO grading to overall survival in a large retrospective cohort of patients with gliomas. We found that CD is a powerful and useful prognostic feature. While WHO grading is still superior, CD information is obtained at far lower cost and risk to the patient.
MATERIALS AND METHODS
Clinical Data
We collected clinical data under a Health Insurance Portability and Accountability Act–compliant retrospective chart review protocol approved by our institutional review board with a waiver of informed consent. Clinical databases were queried for all records of patients diagnosed with gliomas who ultimately underwent surgical resection at our institution. The returned records spanned 1993 to 2018. The resulting clinical data that were analyzed included age, preoperative performance status, surgery dates, imaging dates, follow-up dates, vital status, and diagnoses, including WHO grade. A majority of patients were treated before the introduction of integrated histomolecular diagnoses as introduced in the 2016 revision of the WHO grading system and further emphasized in the 2021 revision.1,2 Therefore, the grades reported are based, for most cases, on morphologic characteristics consistent with the WHO 2007 grading scale. We staged patients on the basis of preoperative imaging data, similar to WHO staging, which is obtained at diagnosis and is not subsequently altered. Thus, the effects of operative, chemotherapy, and radiation therapy treatment were not considered in the current analysis. Overall survival was calculated from the surgery date to the last documented follow-up time, with appropriate right censoring. The patient cohort was further refined by inclusion criteria of 18 years of age or older, WHO grade II, III, or IV gliomas, and the availability of suitable preoperative MR imaging.
Imaging Data
For each patient, preoperative imaging was queried directly from the PACS system. A summary of the sequence parameters for each image type is given in the Online Supplemental Data, and a detailed description of the data processing is provided in the Online Supplemental Data.
Images were skull-stripped to remove nonbrain tissues and coregistered.10,11 Then, tumors were segmented using a pretrained deep learning model, and CSF ROIs were generated using automated Gaussian mixture modeling.12 Additional details of these methods are provided in the Online Supplemental Data. Each image was normalized by mapping modal intensities of healthy brain and CSF to 0 and 1. Note, this is a slightly different scheme than the one used by Gates et al6 but achieves comparable modeling results.
Using the normalized images, we estimated the CD voxelwise throughout the brain of each patient by applying a pretrained random forest model, which has been previously reported.6 This model was trained on imaging and pathology data from 52 image-guided biopsy samples and estimates CD with a root mean square error of 2099 nuclei/mm2 (the total range in the training data was approximately 14,000 nuclei/mm2) using 4 conventional imaging sequences (T1-weighted, T2-weighted, FLAIR, and T1 postcontrast). Examples of the CD maps are shown in Fig 2. As previously reported, these maps agree with literature values for white matter of around 3000 nuclei/mm2 and clinical intuition showing more heterogeneous and highly cellular disease with increasing clinical WHO grade.13 Using these maps, we measured the maximum (max) CD within the visible tumor ROI, defined by the extent of T2/T2-FLAIR hyperintensity, assuming that maximal cellularity is unlikely to occur outside the radiographically visible region. Specifically, we recorded the max CD in the visible lesion after excluding the values in the highest 0.01 cm3 of the measurement ROI as outliers. This process provides a stabilized measure of the maximum that is less sensitive to outliers than the voxelwise maximum. A detailed description is provided in the Online Supplemental Data. For routine clinical use, this measurement could be manually approximated using the mean CD in a small circular ROI of about 10 voxels across (area, about 75 voxels) in the area of highest cellularity. CD maps and the MR imaging data were manually reviewed (by E.D.H.G., with 5 years of experience) using a custom data-review dashboard implemented in R Shiny.14 Studies with unacceptable quality, like failed image registration or excessive artifacts, were excluded from further analysis.
Statistical Analysis
We used Cox proportional hazards modeling and concordance indices (C-indices) to correlate clinical and image features with survival.15,16 We searched for an optimal stratifying threshold in terms of the hazard ratio to create two resulting groups. This procedure was performed within 10-fold cross-validation to prevent overfitting and false discovery of survival differences.17 Statistical differences between the pooled high- and low-risk groups were assessed using a log-rank test and the Kaplan-Meier method.
We performed both univariate and multivariate analysis with adjustments for age and performance status (Karnofsky performance status [KPS]), then again with adjustments for age, KPS, and high WHO grade. Patients who are older and have worse performance status are known to have a poorer prognosis irrespective of other prognostic factors.18,19 We corrected the univariate significance level to account for the number of cellularity measurements tested, using the Benjamini-Hochberg method.20 For simplicity, we report only the best-performing CD feature, max CD. Comparison of the overall correlation between max CD as a predictor (as opposed to a measurement at a single cutoff point) can be accomplished with the C-index, which measures the degree of agreement between a set of predictors and actual survival over the entire curve.21 Comparisons of C-indices from proportional hazards models were performed using jackknife estimates of variance.22 Last, we applied receiver operating characteristic (ROC) analysis with CD measurements and WHO grades to identify a pair of optimal thresholds to separate the WHO grade II, III, and IV tumors using max CD and compared the agreement.
RESULTS
Clinical Data
A summary of the patient cohort selection process is shown in Fig 1. Among 2588 patients whose first resection was at our institution, 1718 had diagnostic imaging available. Of those, 329 had previous biopsies (as opposed to resections), and we elected to include these patients in the analysis. Exclusions were made for pediatric patients or those with WHO grade I (n = 113) and patients with insufficient MR imaging to apply predictive modeling (n = 225). After imaging data review, 199 further cases were excluded for unacceptable data quality. The most common failures were tumor segmentation (5.0% of data) and image registration (3.5% of data). The clinical characteristics of the remaining analyzable 1181 patients are summarized in Table 1.
Flow chart for patient selection in the retrospective data. Ambiguous imaging time means the imaging and operation were on the same day. A complete study includes at least one of each of the following: T1-weighted precontrast, T1-weighted postcontrast, T2-weighted, and FLAIR images.
Clinical data summary of the 1181 cases analyzeda
Correlation between Survival and CD
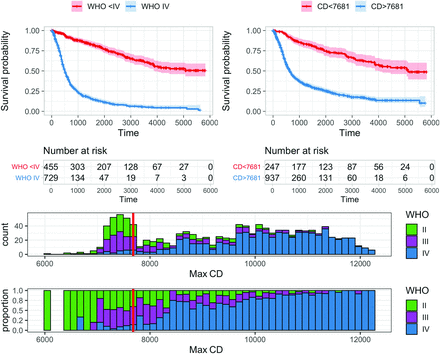
We compared max CD (Fig 2) as a prognostic predictor with WHO grading, with age and KPS as covariates.18,19 The univariate and multivariate hazard ratios are listed in Table 2. Max CD showed the largest survival difference among CD-based features. The optimal threshold of 7681 nuclei/mm2 was very consistent in cross-validation (Online Supplemental Data). Low- and high-risk assignments between cross-validation and in-sample results differed for only 1 patient. The median survival for patients with highly cellular (max CD, >7681 nuclei/mm2) tumors was 630 days compared with 5120 days for patients with low-cellularity tumors. The univariate hazard ratio between the 2 groups was 4.21, adjusted to 2.91 after correcting for covariates of age and KPS (all statistically significant), (Table 2 and Fig 3).
Maps of estimated CD for gliomas of known WHO grades. T2-weighted images are shown in the upper row for reference. Note the graphic nature of CD estimates and how CD maps can be used for image-guided therapy.
Survival modeling for patients with all WHO grades (II, III, IV)a
The best CD (in nuclei/square millimeters) measure for dichotomizing survival in adult gliomas is a max CD (stabilized with 0.01-cm3 volume constraint) of 7681 nuclei/mm2. Upper left: Patients with WHO grade IV gliomas have much worse overall survival (median, 497 days) than patients with WHO II or III gliomas (median unreached, 75% survival at 2131 days). Upper right: An optimal cutoff (maximizing hazard ratio) of 7681 nuclei/mm2 divides the glioma population into groups with median survivals of 630 and 5120 days, respectively (log-rank, P < .001). Lower row: A histogram of all cases (bin size = 123 nuclei/mm2) demonstrates stabilized max CD values ranging from 6089 to 12,260 with an interquartile range of 7632–10,717 and a mean of 9175. WHO grade II, III, and IV cases are color-coded, and the optimized cutoff value used in the upper right panel (7681 nuclei/mm2) is shown with a solid red line. The lower histogram has all bins scaled to height 1 to show the relative proportion of each WHO grade in each bin.
For comparison, the hazard ratio for histologically defined WHO grade IV disease was 7.57 on univariate analysis relative to WHO grade II and III, decreased to 5.25 for multivariate analysis when correcting for age and KPS. Max CD had C-indices of 0.662 alone, and 0.734 after adjusting for covariates, which compared well with WHO grading at 0.704, and 0.761 after adjusting for covariates (Online Supplemental Data). The concordance indices were significantly different from each other before (P < .001) and after (P < .001) adjusting for covariates.
In a combined model with age, KPS, WHO grade IV, and max CD analyzed together, the multivariate hazard ratio for CD was 1.36 (P < .05). The effect of WHO grade (hazard ratio = 4.60) was still larger, however, in the same model (Table 2). This combined model gave a risk score with a C-index of 0.764 with overall survival (95% CI, 0.748–0.781). This was significantly higher than the C-index for the model using just age, KPS, and a WHO grade of 0.761 (P = .002) (Online Supplemental Data). Again, this finding suggests some overlap but with nonredundant information present between WHO grading and max CD.
Correlation between CD and WHO Grade
The histogram of max CD values in Fig 3 shows a striking relationship between tumors with a high WHO grade (WHO IV) and larger maximum cellularity. The optimal threshold with respect to survival of 7681 nuclei/mm2 effectively divides the high-grade (WHO grade IV) from the low-grade (WHO grade II and III) cases with a 93% sensitivity. Max CD showed no ability to differentiate WHO II from WHO III tumors due to the high overlap in the histograms. However, we were able to construct a trio of risk categories using CD that mimics the WHO II, III, and IV risk stratification, (Fig 4). We selected 2 cutoff points at 7443 and 8358 nuclei/mm2 via ROC analysis to optimally mimic the WHO groups. These values are different from the previously mentioned 7681 nuclei/mm2 cutoff, which was chosen to optimize overall survival differences between just two groups of patients. The number of patients and median survival for each group are tabulated in Table 3. These resulting three categories showed risk stratification visually similar to the WHO grades, though there were statistical differences in median overall survival (log-rank, P = .004).
Upper left: Survival curves stratified by WHO grade. Median survival for WHO grades II, III, and IV: unreached, 3486 days, 497 days, respectively. Upper right: CD thresholds at 7443 and 8358 nuclei separate patients into 3 survival groups that are similar to the WHO II, III, and IV groups (median survival unreached, 3738 days, and 603 days, respectively). There is a significant difference in the red and blue survival curves (log-rank, P = .004). Lower right: P-spline fit (3 degrees of freedom) for nonlinear estimation of the hazard ratio with respect to CD for constant age and KPS. The reference point, hazard ratio 1, is arbitrary. The shaded background corresponds to the three groupings based on CD shown in the upper right panel. Dashed lines are 95% confidence intervals. Relative risk increases with increasing CD up to a max CD of about 9000 nuclei/mm2, after which it plateaus. Max CD allows a continuous estimate of risk, unlike the categoric WHO grade.
Patient numbers by max CD ranges (nuclei/square millimeter) and WHO gradea
One advantage of CD as a risk measure over WHO grading is that the estimated CD is a continuous measurement that can provide finer risk-stratification groups than the three-class categoric WHO grade (WHO I disease was not found in our adult population with gliomas, reducing the analysis to three categories). In proportional hazards modeling, the relation between a continuous measurement like CD and the hazard ratio is assumed to be log-linear. However, a nonlinear fit can be achieved using spline fitting. Figure 4 also shows the resulting nonlinear fit with the grade-matched cutoffs overlaid. The plateau at higher CD values (visually about >9000 nuclei/mm2) suggests a saturation-type effect beyond which increased max CD does not further increase risk. At lower CD values, the curve is steeper (ie, greater sensitivity of risk to CD changes), suggesting that CD might allow more precise risk stratification for lower-grade gliomas. The nonlinear spline fit illustrates the relation between CD and risk at various CD levels but does not significantly improve concordance of the Cox model (C-index difference, 8 × 10−4; P = .65).
DISCUSSION
We estimated CD using MR imaging and a machine learning algorithm in a retrospective cohort of 1181 previously untreated patients with gliomas from our institution. We found that high max CD indicates worse prognosis independent of age, performance status, and even WHO tumor grade. The prognostic power of max CD is slightly less than that of the WHO grade but comes remarkably close, especially given the relatively low cost and risk of obtaining these CD estimates. The difference in concordance between a model based on WHO grade (and covariates) and the model based on CD (and covariates) was just 0.027 (95% CI, 0.016–0.037). CD estimates also have advantages over WHO grading, including timeliness, lower risk, and lower cost of the estimates. The graphic nature of the estimates also allows CD estimates to be used for image-guided therapies.
CD is known from the literature to correlate to survival.23 Conventional and physiologic techniques like T2-FLAIR or DWI correlate with increased tissue cellularity.24,25 Recently, several research studies have used machine learning trained on MR imaging and tissue data to quantitatively estimate CD in gliomas from imaging data alone. These models produce graphic mapping of CD that characterizes the full tumor heterogeneity and shows promising clinical applications such as identifying hypercellular regions outside contrast enhancement.6-9 Our study differs from the current literature in that we directly evaluated the correlation between measures of cellularity and survival outcome. We focused specifically on simple, interpretable, first-order measures of cellularity rather than complex nonlinear feature combinations like texture analysis or deep filter features. Estimated cellularity maps already combine multiple sources of information from the MR images and tissue-training data, possibly rendering additional complexity unnecessary. Another key difference of our study is that we use a combined cohort of multiple WHO grades to correlate with survival, mimicking the actuality of practice before tissue diagnosis is known.
One limitation of this work is gaps in the clinical data for the retrospective cohort. We account for overall survival of patients with different WHO-grade gliomas, ranging from about 12 months (WHO IV glioblastoma) to >5 years (WHO II).26,27 However, we were not able to include therapeutic intervention or mutational profiles, which also affect outcomes.18,27-29 Survival differences due to chemoradiation or total tumor resection range from a few months to more than a year,27,30 and IDH1 mutation is associated with a nearly 2-fold difference in median overall survival.31 Most of the patients in our cohort were treated before 2015, when molecular information started being routinely collected.
Related to imaging, the retrospective nature of our study limited our control over specific imaging sequences. This issue caused some inaccuracy in the random forest that estimated cellularity because the forest was trained on data from a tightly controlled research protocol.6 Intensity normalization accounts for much of the variability in image contrast and acquisition parameters, but the true accuracy on the retrospective data cannot be known without extensive histologic sample verification. Although the random forest estimates of CD using the conventional sequences have only a moderate correlation to actual measured CD (R2 = 0.52),6 survival models based on these estimates achieved a high concordance (C = 0.73) with overall survival. Two factors can explain this: First, the continuous CD estimates are binned with a threshold to designate high-risk and low-risk patients, generally an easier task than precise quantitative estimation. Second, clinical factors like age and KPS aid survival models. The fact that even rough estimates of cellularity are effective in estimating survival reinforces their potential value.
Additionally, DWI was not commonly available in the historic patient cohort. Given the well-established relationship between CD and DWI, we are eager to include this as part of future analyses. Finally, we examined the effect of preoperative cellularity on prognosis without explicitly accounting for treatment variables like gross total–versus-subtotal surgical resection or radiation treatment. While these are important, quantifying changes in CD after therapy is a very challenging image-processing task and is the subject of future investigation.
For future work, one of the most valuable directions is more accurately modeling cellularity in normal tissue like gray matter and white matter. Expanded training data in the random forest for these anatomic areas is a possible solution. Another potential solution is to apply methods from MR image synthesis to supplement the random forest to generate high-quality CD mappings (such as in Fig 2).32 Graphical maps, either in their present form or future improved versions, will enable prospective clinical trials to validate the accuracy of cellular density estimates.
CONCLUSIONS
We evaluated the correlation between estimated glioma cellularity and survival in a large retrospective cohort of adults with infiltrative gliomas. We showed that imaging estimates of CD are a powerful and independent prognostic predictor of survival, only slightly inferior to gold standard WHO grading. CD estimates are useful as a supplement to WHO grading due to the information being 1) timelier and available before any surgical procedures, 2) less risky to obtain than an open procedure, and 3) less costly than tissue sampling. CD estimates are useful beyond prognosis estimation in that graphic maps of CD can be used to guide biopsy and reduce the risk of undergrading and should be helpful in planning surgery and radiation treatment. In addition, CD as a continuous variable might allow more precise risk stratification, compared with the relatively coarse granularity of the categoric WHO grading scheme. Future clinical trials to prospectively test imaging-based CD estimates are justified.
Footnotes
Data for this work have been obtained through a search of the integrated multidisciplinary Brain and Spine Center Database. The Brain and Spine Center Database was supported, in part, by an institutional MD Anderson database development grant. The High-Performance Research Computing facility at the University of Texas MD Anderson Cancer Center provided computational resources that have contributed to the research results reported in this work.
E.D.H. Gates was supported by a training fellowship from the Gulf Coast Consortia on the NLM Training Program in Biomedical Informatics & Data Science (T15LM007093). D. Fuentes, D. Schellingerhout, and A. Celaya were partially supported by R21CA249373. J.P. Long was partially supported by the National Cancer Institute and the National Center for Advancing Translational Sciences of the National Institutes of Health [P30CA016672 and CCTS UL1TR003167].
Disclosure forms provided by the authors are available with the full text and PDF of this article at www.ajnr.org.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received February 10, 2022.
- Accepted after revision July 1, 2022.
- © 2022 by American Journal of Neuroradiology